Team:IvyTech SouthBend IN/progress
From 2014.igem.org
(Difference between revisions)
| Line 69: | Line 69: | ||
<li><a href="#929104">9/29/14-10/4/14</a></li> | <li><a href="#929104">9/29/14-10/4/14</a></li> | ||
</ul> | </ul> | ||
| + | <!-- column 2 end --> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="ivymask leftmenu"> | ||
| + | |||
| + | <div class="ivyleft"> | ||
| + | <div class="ivy1"> | ||
| + | <!-- column 1 start --><br /> | ||
| + | <div id="memo2g"><a name="92128">Week</a> of 10/05/14 through 10/11/14</div> | ||
| + | <ul> | ||
| + | <li>This week we began work on the nanotechnology aspects of our project. The main focus was working on the microfluidic channel, the volume of space that is going to hod the water sample while the biochemical reaction for beta-galactosidase is taking place. We used negative lithography to cast a master mold of a chamber we designed and thought would be best suited for our device. After the mold was cast and sent through numerous bake sessions, it was inspected and determined to be an acceptable mold. We then made a polydimethylsiloxane (PDMS) mixture with a 10:1 ratio with a curing agent. After pouring it into the mold, great care was taken to ensure there were no bubbles left in the PDMS mixture. After being left to dry and hardened, a plasma generator was used to bind the cut out piece to a glass slide. Unfortunately, after testing it with ink we got negative results. Two chambers were tested and both collapsed. We are going to try again using shorter channels and combining pillars so there is a better support system. </li><br /> | ||
| + | <li>We also ran a protocol of various monolayers to test adhesion for the most favorable surface for biofilm adhesion. We are doing this to make a more optimal environment for the lyopholized cells that will be kept in the chamber of our device until a sample is introduced. 2% solution in acetone was used. After monolayers were added to slides they were incubated while being soaked in broth cultures of Top 10 E. coli cells. (3-Aminopropyl)triethoxysilane (APTES) was found to have the most cells attached when counting results under a microscope. </li> | ||
| + | </ul> | ||
| + | <!-- column 1 end --> | ||
| + | </div> | ||
| + | <div class="ivy2"> | ||
| + | <!-- column 2 start --> | ||
| + | <p><center><img src="http://i.imgur.com/0Oyp5dj.png" style="width: 75%;max-height: 75%" /></center></p> | ||
<!-- column 2 end --> | <!-- column 2 end --> | ||
</div> | </div> | ||
| Line 81: | Line 101: | ||
<div id="memo3g"><a name="929104">Week</a> of 9/29/14 through 10/4/14</div><br /> | <div id="memo3g"><a name="929104">Week</a> of 9/29/14 through 10/4/14</div><br /> | ||
<ul> | <ul> | ||
| - | <li>This week our team had to split up. Two of our members-Kevin and Lyuda- went along with our adviser Professor Twaddle to Posters on the Hill for CCURI (Community College Undergraduate Research Initiative) in | + | <li>This week our team had to split up. Two of our members-Kevin and Lyuda- went along with our adviser Professor Twaddle to Posters on the Hill for CCURI (Community College Undergraduate Research Initiative) in Washington D.C. While they were away, Karinne worked on subculturing into fresh broth so that we can make sure to have clean cell lines to work with.</li><br /> |
<li>Also, after an exchange of emails with the Carnegie Melon iGEM team we realized we would be able to help test a beta kit of theirs that make the DNA of wheat germ visible after only a few simple steps. It was easy to use and easy to understand and we thank the team profusely for the fun experience of of this experiment!</li> | <li>Also, after an exchange of emails with the Carnegie Melon iGEM team we realized we would be able to help test a beta kit of theirs that make the DNA of wheat germ visible after only a few simple steps. It was easy to use and easy to understand and we thank the team profusely for the fun experience of of this experiment!</li> | ||
</ul> | </ul> | ||
| Line 101: | Line 121: | ||
<div id="memo2g"><a name="92128">Week</a> of 9/21/14 through 9/28/14</div> | <div id="memo2g"><a name="92128">Week</a> of 9/21/14 through 9/28/14</div> | ||
<ul> | <ul> | ||
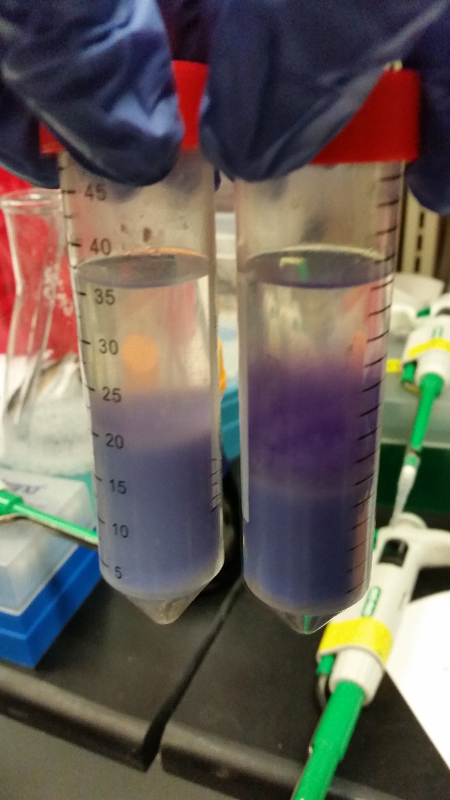
| - | <li>This week we are going to keep working on our assay of Beta-galactosidase activity. So far we have had to unsuccessful attempts at our run-through of it with intact cells. This week we started it a third and fourth time and had success! Our E. coli C turned to a red color along with our K1477014 part. Not only that, but our negative control stayed negative. Everything worked out perfectly except for the time elapsed to get these results. We expected it to take 12-14 hours for a color change to occur, but it ended up taking about 36 hours in one instance and a little over 48 hours in the last attempt. However we are still pleased to have | + | <li>This week we are going to keep working on our assay of Beta-galactosidase activity. So far we have had to unsuccessful attempts at our run-through of it with intact cells. This week we started it a third and fourth time and had success! Our E. coli C turned to a red color along with our K1477014 part. Not only that, but our negative control stayed negative. Everything worked out perfectly except for the time elapsed to get these results. We expected it to take 12-14 hours for a color change to occur, but it ended up taking about 36 hours in one instance and a little over 48 hours in the last attempt. However we are still pleased to have achieved success (we are still fighting contamination.)</li><br /> |
| - | <li>We also did two runs of this assay with chemically lysed cells. In the first run through, we got positive results. However, an inquiry led to the discovery that one of the chemicals used could potentially denature any enzymes present, and so we had to throw out that data. The second time we used an | + | <li>We also did two runs of this assay with chemically lysed cells. In the first run through, we got positive results. However, an inquiry led to the discovery that one of the chemicals used could potentially denature any enzymes present, and so we had to throw out that data. The second time we used an appropriate chemical, but had positive results on everything-including the negative control. We are now out of our required materials for chemically lysing cells.</li> |
</ul> | </ul> | ||
<!-- column 1 end --> | <!-- column 1 end --> | ||
| Line 121: | Line 141: | ||
<div id="memo3g"><a name="91420">Week</a> of 9/14/14 through 9/20/14</div><br /> | <div id="memo3g"><a name="91420">Week</a> of 9/14/14 through 9/20/14</div><br /> | ||
<ul> | <ul> | ||
| - | <li>This week we began our attempts on the Beta-galactosidase | + | <li>This week we began our attempts on the Beta-galactosidase activity assay. We plan to run this assay three different ways. We will first complete it using intact cells, measuring the activity that is produce inside the cell. Then we will use chemically lysed cells. By lysing the cells in this way, we are ensuring a higher accuracy in regards to amount of cells being lysed. The chemicals will destroy the cell wall and the beta-gal complimentation can occur in vitro.</li><br /> |
<li>Finally, we plan to bring the core idea of our research project together, and then we will run this assay using cells that have been lysed with bacteriophage.</li><br /> | <li>Finally, we plan to bring the core idea of our research project together, and then we will run this assay using cells that have been lysed with bacteriophage.</li><br /> | ||
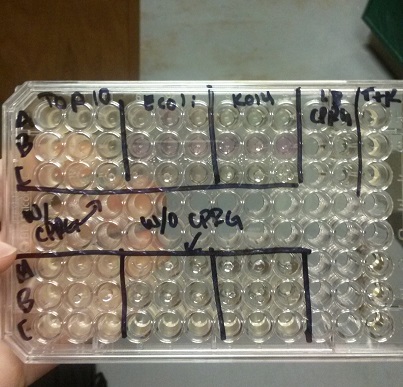
| - | <li>In all of these, we will be using chlorophenol red (CPRG) as the substrate. This will cause a color change that ranges from yellow to dark red. We are using Top 10 E. coli cells ad our negative control, as these only produce one out of the two parts that make up the enzyme. We will be using E. coli C as our positive control. We are testing the | + | <li>In all of these, we will be using chlorophenol red (CPRG) as the substrate. This will cause a color change that ranges from yellow to dark red. We are using Top 10 E. coli cells ad our negative control, as these only produce one out of the two parts that make up the enzyme. We will be using E. coli C as our positive control. We are testing the activity produced in our part K1477014 transformed into Top 10 cells.</li><br /> |
<li>Our first try did not yield correct results, with all three cell samples leading to a positive color change. Our second attempt also did not turn out right. Only the E. coli C gave a positive reading of Beta-gal activity.</li> | <li>Our first try did not yield correct results, with all three cell samples leading to a positive color change. Our second attempt also did not turn out right. Only the E. coli C gave a positive reading of Beta-gal activity.</li> | ||
</ul> | </ul> | ||
| Line 204: | Line 224: | ||
<div id="memo2g"><a name="82430">Week</a> of 8/24/14 through 8/30/14</div> | <div id="memo2g"><a name="82430">Week</a> of 8/24/14 through 8/30/14</div> | ||
<ul> | <ul> | ||
| - | <li>This week our team's | + | <li>This week our team's persistence and effort came to fruition when we finally achieved a virus titer plaque assay that turned out perfectly! There was no contamination. The cell growth did not overpower the working virus bacteriophage. Best of all, we have clear dead zones that show consistency with the corresponding viral dilutions. We are now ready to begin performing the Beta-galactosidase activity assay.</li> |
</ul> | </ul> | ||
<!-- column 1 end --> | <!-- column 1 end --> | ||
| Line 245: | Line 265: | ||
<ul> | <ul> | ||
<li>During this week we restocked our supplies for broth and agar growth media, as we were running very low. </li><br /> | <li>During this week we restocked our supplies for broth and agar growth media, as we were running very low. </li><br /> | ||
| - | <li>One of our | + | <li>One of our advisers suggested we change the buffer we were using for our gel electrophorisis, and that when preparing the DNA mixture to insert into the gel, we should use buffer instead of DDI water, since this is what is present when the gel is running.</li><br /> |
<li>However, even after changing the buffer, we did not experience any better results. We tried a few times. First, the DNA of our parts did not show up at all. And again, our DNA ladder did not show up correctly.</li> | <li>However, even after changing the buffer, we did not experience any better results. We tried a few times. First, the DNA of our parts did not show up at all. And again, our DNA ladder did not show up correctly.</li> | ||
</ul> | </ul> | ||
| Line 263: | Line 283: | ||
<div id="memo3g"><a name="839">Week</a> of 8/3/14 through 8/9/14</div><br /> | <div id="memo3g"><a name="839">Week</a> of 8/3/14 through 8/9/14</div><br /> | ||
<ul> | <ul> | ||
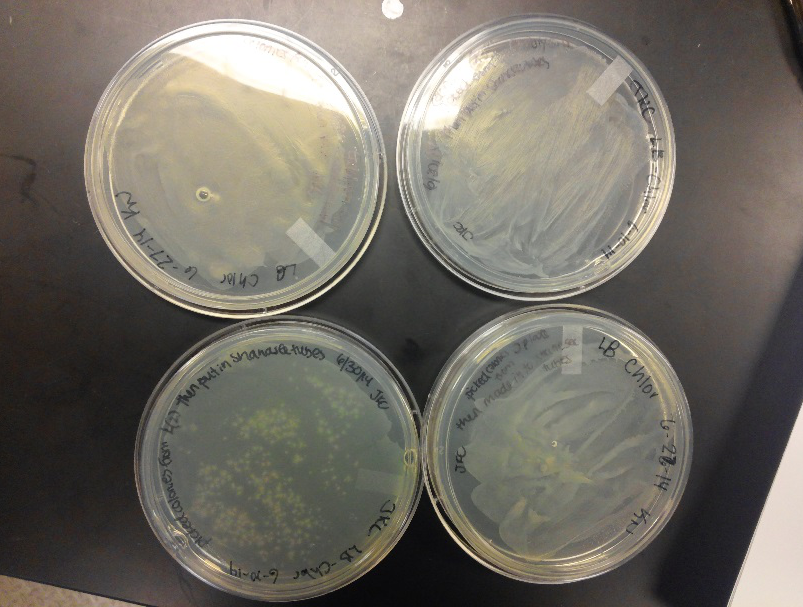
| - | <li>During this week, the team realized that the contamination was coming from our stock of cells that was being kept in the -80C freezer. We have no idea how they became contaminated, seeing as continued to use the line of cells we froze for a week after we froze them, but before any contamination occurred. Nonetheless, one of our | + | <li>During this week, the team realized that the contamination was coming from our stock of cells that was being kept in the -80C freezer. We have no idea how they became contaminated, seeing as continued to use the line of cells we froze for a week after we froze them, but before any contamination occurred. Nonetheless, one of our advisers has procured a new stock of top 10 E. coli cells for us to use.</li><br /> |
<li>After unsuccessful runs of electrophoresis gels, we have run out of DNA to try to measure. So, we took some time out this week to streak, grow, and purify lines of our K1477014 and K1477030 parts on both pSB1C3 and pSB1AT3 backbones.</li> | <li>After unsuccessful runs of electrophoresis gels, we have run out of DNA to try to measure. So, we took some time out this week to streak, grow, and purify lines of our K1477014 and K1477030 parts on both pSB1C3 and pSB1AT3 backbones.</li> | ||
</ul> | </ul> | ||
| Line 277: | Line 297: | ||
<div id="memo2g"><a name="72782">Week</a> of 7/27/14 through 8/2/14</div> | <div id="memo2g"><a name="72782">Week</a> of 7/27/14 through 8/2/14</div> | ||
<ul> | <ul> | ||
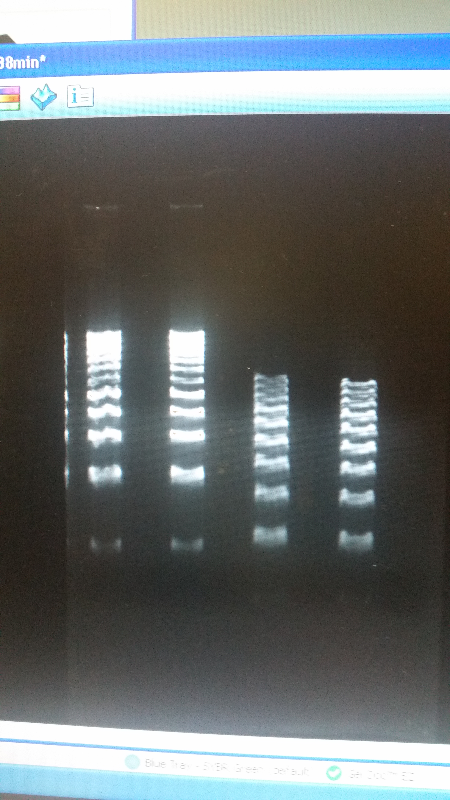
| - | <li>After talking with an outside | + | <li>After talking with an outside adviser, we have realized that much of our protocol was wrong when it came to running an electrophoresis gel. We were doing a "quick run" protocol, running a high voltage for a short time. Now we are running at a significantly lower voltage, and tripling the time it takes to complete the gel. The DNA bands to our parts are now turning out well. However, the bands in the DNA ladder still look smashed together, which continues to make it impossible to count our parts' kilobases.</li><br /> |
<li>Our team has started working on a plaquing assay with T7 and T4 viruses. We are trying to figure out exactly what concentration of virus we need to work with. Unfortunately, the problem of contamination has become an even bigger issue during the performance of this protocol. The outcome yields a lawn growth of cells that is too thick to make out sufficient plaques of dead zones and there are bright yellow colonies found on a few of the plates (Five plates total in all.) We worry also that perhaps we are using a broth culture of top 10 E.coli cells whose concentration may be too high for the virus to kill.</li> | <li>Our team has started working on a plaquing assay with T7 and T4 viruses. We are trying to figure out exactly what concentration of virus we need to work with. Unfortunately, the problem of contamination has become an even bigger issue during the performance of this protocol. The outcome yields a lawn growth of cells that is too thick to make out sufficient plaques of dead zones and there are bright yellow colonies found on a few of the plates (Five plates total in all.) We worry also that perhaps we are using a broth culture of top 10 E.coli cells whose concentration may be too high for the virus to kill.</li> | ||
</ul> | </ul> | ||
| Line 297: | Line 317: | ||
<div id="memo3g"><a name="72026">Week</a> of 7/20/14 through 7/26/14</div><br /> | <div id="memo3g"><a name="72026">Week</a> of 7/20/14 through 7/26/14</div><br /> | ||
<ul> | <ul> | ||
| - | <li>We have created a stock of our two parts on the pSB1C3 backbone. We purified both parts will prepare to send them in to iGEM by running am electrophoresis gel to prove that our parts are what we say they are. We ran two gels, but the DNA bands are coming out deformed for some reason and the DNA ladder does not have | + | <li>We have created a stock of our two parts on the pSB1C3 backbone. We purified both parts will prepare to send them in to iGEM by running am electrophoresis gel to prove that our parts are what we say they are. We ran two gels, but the DNA bands are coming out deformed for some reason and the DNA ladder does not have distinguished bands that can be used to count how many kilobases our part has. We are looking into why this is. Possible explanations include incorrect voltage, incorrect agarose gel thickness, or incorrect amount of DNA added. Adjustments will be made in the future.</li><br /> |
| - | <li>We have also | + | <li>We have also performed biobirck assembly two more times and have a new stock of our part on its original backbone pSB1AT3.</li><br /> |
<li>Our problem of contamination is continuing to persist. During our attempts of recreating a new stock of our parts, we found bright yellow colonies all over more than half of the plates we streaked.</li> | <li>Our problem of contamination is continuing to persist. During our attempts of recreating a new stock of our parts, we found bright yellow colonies all over more than half of the plates we streaked.</li> | ||
</ul> | </ul> | ||
| Line 319: | Line 339: | ||
<ul> | <ul> | ||
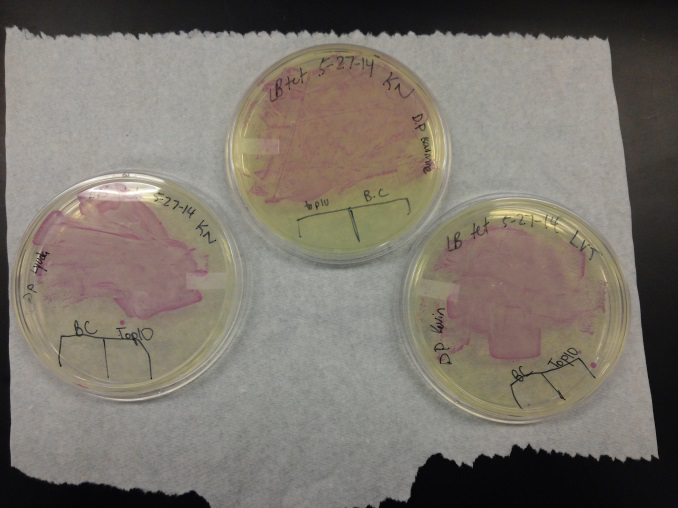
<li>This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.</li><br /> | <li>This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.</li><br /> | ||
| - | <li>Unfortunately, We no longer have any stock of our original part. So, now we are again following the Biolabs Biobrick Assembly Manual to | + | <li>Unfortunately, We no longer have any stock of our original part. So, now we are again following the Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.</li> <br /> |
<li>Our second run through of the protocol resulted in the in the growth of red colonies on chlor plates after electroportion of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.</li> | <li>Our second run through of the protocol resulted in the in the growth of red colonies on chlor plates after electroportion of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.</li> | ||
</ul> | </ul> | ||
| Line 334: | Line 354: | ||
<ul> | <ul> | ||
<li>Last week Lyuda and Kevin ligated the composite part from the pSB1AT3, to the pSB1C3. We had difficulty getting the plasmid backbone to take the transformation. We then emailed the IGEM FAQ section and got an almost immediate response stating that the chlor backbone had a really low transformation efficiency. We tried again on 7-20 the ligation of both K1477030, and K1477014 with success but it takes like two days in the incubator.</li><br /> | <li>Last week Lyuda and Kevin ligated the composite part from the pSB1AT3, to the pSB1C3. We had difficulty getting the plasmid backbone to take the transformation. We then emailed the IGEM FAQ section and got an almost immediate response stating that the chlor backbone had a really low transformation efficiency. We tried again on 7-20 the ligation of both K1477030, and K1477014 with success but it takes like two days in the incubator.</li><br /> | ||
| - | <li>We are in the process of running a gel to prove our parts are the right size in | + | <li>We are in the process of running a gel to prove our parts are the right size in base pairs to show proper replacement transformations.</li><br /> |
<li>We have resuspended our composite parts in LB and the appropriate antibiotics to begin mass producing for concentration testing. We are awaiting the arrival of viruses for furthering our virus titering protocol. | <li>We have resuspended our composite parts in LB and the appropriate antibiotics to begin mass producing for concentration testing. We are awaiting the arrival of viruses for furthering our virus titering protocol. | ||
We now have duplicate lines of all composite parts growing and in suspension all prepared for moving into concentration testing. PROGRESS!!</li><br /> | We now have duplicate lines of all composite parts growing and in suspension all prepared for moving into concentration testing. PROGRESS!!</li><br /> | ||
Revision as of 15:13, 13 October 2014
Week of 8/3/14 through 8/9/14
- During this week, the team realized that the contamination was coming from our stock of cells that was being kept in the -80C freezer. We have no idea how they became contaminated, seeing as continued to use the line of cells we froze for a week after we froze them, but before any contamination occurred. Nonetheless, one of our advisers has procured a new stock of top 10 E. coli cells for us to use.
- After unsuccessful runs of electrophoresis gels, we have run out of DNA to try to measure. So, we took some time out this week to streak, grow, and purify lines of our K1477014 and K1477030 parts on both pSB1C3 and pSB1AT3 backbones.
Week of 7/13/14 through 7/19/14
- This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.
- Unfortunately, We no longer have any stock of our original part. So, now we are again following the Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.
- Our second run through of the protocol resulted in the in the growth of red colonies on chlor plates after electroportion of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.
Week of 5/25/14 through 5/31/14
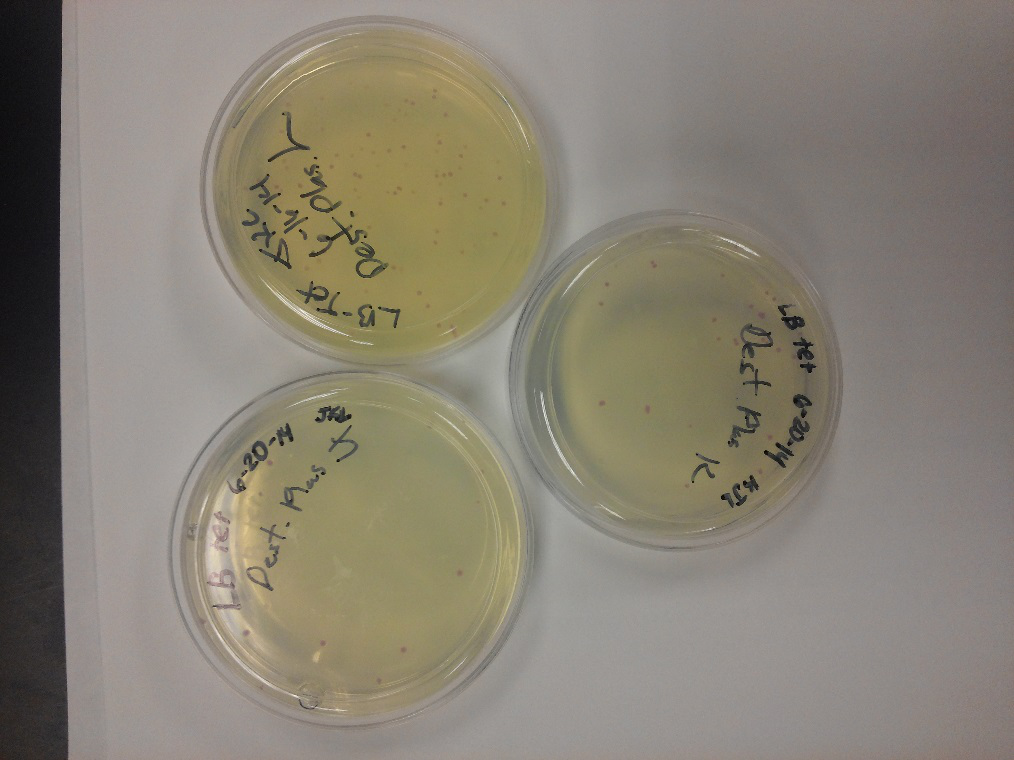
- We transformed Top 10 cells through electroporation with registry parts K112806 (Endolysin,) I732020 (LacZoperon Mutant,) J23102 (p-con) and I712074 (T7 promoter.)
- After streaking on plates, and leaving them in the incubator for a day, there was growth on all plates, showing that a successful transformation occured.
- Plates containing J23102 were red-another sign of success. Unfortunately, it looks like contamination has arisen as well, and is spreading on plates with I712074 and K112806 growth.
- The contamination persisted all week, but we were able to get plates with transformed parts that contained no contamination growth.
- We then ran a DNA purification protocol of J23102 and I732020 to get them ready for a biobrick assembly protocol.
 "
"