Team:SUSTC-Shenzhen/Modeling/Interlab
From 2014.igem.org
Zhangysh1995 (Talk | contribs) |
Zhangysh1995 (Talk | contribs) |
||
| Line 93: | Line 93: | ||
===='''Results'''==== | ===='''Results'''==== | ||
| + | <html><div class="table-responsive"></html> | ||
| + | {|class="table" | ||
| + | ! Position | ||
| + | ! A | ||
| + | ! B | ||
| + | ! C | ||
| + | ! D | ||
| + | ! E | ||
| + | ! F | ||
| + | ! G | ||
| + | ! H | ||
| + | |- | ||
| + | ! 1 | ||
| + | | J23101_Kana | ||
| + | | J23101_Kana | ||
| + | | J23101_Kana | ||
| + | | J23101_GFP | ||
| + | | J23101_GFP | ||
| + | | J23101_GFP | ||
| + | | - | ||
| + | | - | ||
| + | |- | ||
| + | !2 | ||
| + | | J23115_Kana | ||
| + | | J23115_Kana | ||
| + | | J23115_Kana | ||
| + | | J23115_GFP | ||
| + | | J23115_GFP | ||
| + | | J23115_GFP | ||
| + | | - | ||
| + | | - | ||
| + | |- | ||
| + | !3 | ||
| + | | I20260_Kana | ||
| + | | I20260_Kana | ||
| + | | I20260_Kana | ||
| + | | I20260_GFP | ||
| + | | I20260_GFP | ||
| + | | I20260_GFP | ||
| + | | - | ||
| + | | - | ||
| + | |} | ||
| + | <html></div></html> | ||
| + | Table 1.1 Correspondence | ||
| + | |||
{{SUSTC-Image|wiki/images/a/a4/Amplification_Plot_Rn_vs_Cycle.jpg|qPCR}} | {{SUSTC-Image|wiki/images/a/a4/Amplification_Plot_Rn_vs_Cycle.jpg|qPCR}} | ||
Fig 1.1 Amplification plot for qPCR | Fig 1.1 Amplification plot for qPCR | ||
Revision as of 11:56, 6 October 2014
Inter-lab Study
Methods to test Fluorescence Density
Contents |
What is Inter-lab Study?
Inter-lab study is part of a new tack called Measurement this year. All team are encouraged to participate in it. See more details HERE(https://2014.igem.org/Tracks/Measurement/Interlab_study).
The aim of what we did is to get fluorescence statistics. To achieve this, we did four different sets of experiments:
- Total RNA extraction, reverse transcription and qPCR; (From transcription level)
- Measure with Fluorescence microscope and Image-Pro Plus 6.0 (IPP 6.0); (From protein expression level)
- Measure with Flow cytometry; (From protein expression level)
- Measuren with fluorescence spectrophotometer (From protein expression level)
Plasmid Construction
- 3A assembly was used to construct BBa_J23101 + BBa_E0240 and BBa_J23115 + BBa_E0240. Construction was completed on 9th, Aug. And samples were sent to be sequenced.
- We also tried to construction in Extra Credit Opportunity. But results of electrophoresis with BBa_J04450+BBa_J23101/J23115 show anther segments which we cannot figure out what it was until now.
Measurement
Experiment I: Total RNA extraction, reverse transcription and qPCR
Introduction
OD (Optical density) is the density of light absorbed by the E. coli culture, and it is directly proportional to culture density. We calculated culture volumes according to ratio of OD. After that we did reverse transcription and qPCR to identify mRNA of GFP in each sample for three replicates.
Materials
StepOnePlus™ Real-Time PCR System (Life Technologies), E.Z.N.A.® Bacterial RNA Kit ( Omega bio-tek), GoScript™ Reverse Transcription System (Promega), SYBR® Select Mater Mix (Life Technologies)
Protocols
- RNA extration was done following instruction in the kit. Final concentration:
| Sample name | I20260 | J23101 | J23115 |
|---|---|---|---|
| RNA concentration | 476.4ng/uL | 456.6ng/uL | 446.0ng/uL |
- Reverse Transcription
Step1:
| Sample name | I20260 | J23101 | J23115 |
|---|---|---|---|
| RNA | 2.10uL | 2.19uL | 2.24uL |
| Random primer | 1.00uL | 1.00uL | 1.00uL |
| dd Water | 1.90uL | 1.81uL | 1.76uL |
| Total | 5.00uL | 5.00uL | 5.00uL |
Incubated at 70℃ for 5 minutes, immediately put on ice for 5min, then centrifuge for 30 seconds, at last put on ice.
Step2: Using promega GoScriptTM Reverse Transcription System, incubated at 25℃ for 5 minues, then incubated at 42℃ at 60min, again at 70℃ for 15 minutes.
- qPCR
Holding Stage: 50.0℃ for 2min, 95℃ for 2min.
Cycling Stage: 95℃ for 15sec, 55.0℃ for 15sec, 72.0℃ for 1 min; 40 cycles.
Melt Curve Stage: 60.0℃ for 1min, 95℃ for 15sec.
- The gene coding Kanamycin is recognized as reference gene which is run in the qPCR.
- All samples are run for three replicates.
Results
| Position | A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|---|
| 1 | J23101_Kana | J23101_Kana | J23101_Kana | J23101_GFP | J23101_GFP | J23101_GFP | - | - |
| 2 | J23115_Kana | J23115_Kana | J23115_Kana | J23115_GFP | J23115_GFP | J23115_GFP | - | - |
| 3 | I20260_Kana | I20260_Kana | I20260_Kana | I20260_GFP | I20260_GFP | I20260_GFP | - | - |
Table 1.1 Correspondence
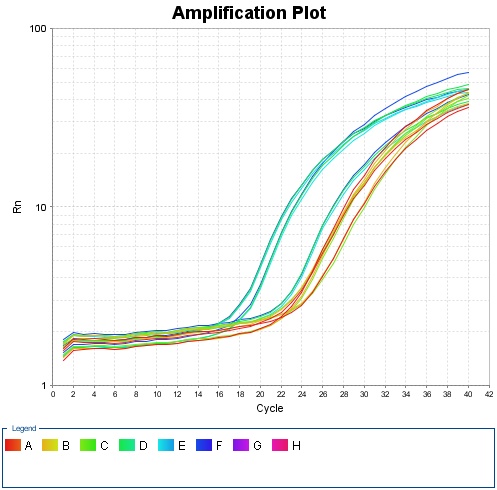
Fig 1.1 Amplification plot for qPCR
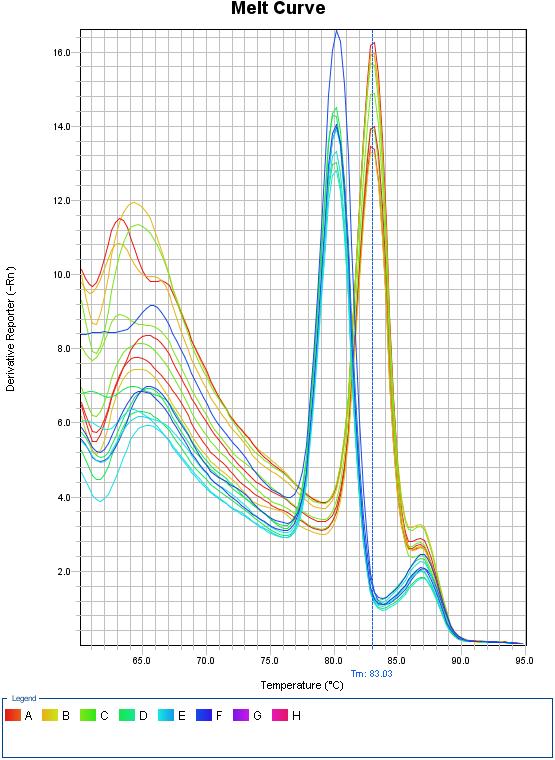
Fig 1.2 Melt curve for qPCR
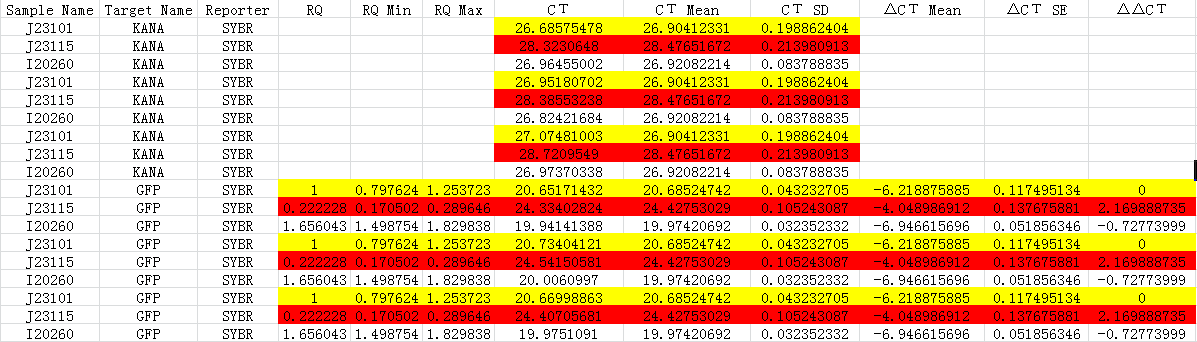
Fig 1.3 Ct for qPCR
Analysis
We took an experiment to find the relation between OD600 and bacterial number and knew that if OD600 is less than 1, OD600 is about a linear relation with the number of bacterial.
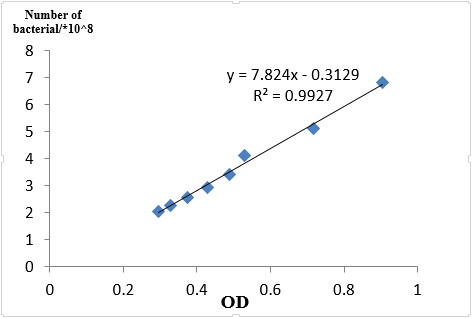
Fig 1.4 OD and Cell number
In case of different growth statues of bacterial, we reshaked bacterial according to 1:100 of germ solution. Finally, the OD600 of J23101 =0.436, J23115 =0.511, I20260=0.560.So when we get the OD600 of each sample, we made the same number of bacterial to extract RNA, J23101 = 3.00ml, J23115 =2.56ml, I20260=2.34ml, all the number in culture is about 3.09*108.
We made J23101 as the control group, 2-ΔΔCT to calculate the relative amount of mRNA.
Equation 1: X=X0(1+Ex)n
X: numbers of target gene after n turn of cycle
X0: initial number of target gene
Ex: amplification efficiency of target gene
n: Cylcles
Equation 2: Xt=X0(1+Ex)Ct,x =Kx
Xt: number of target gene reaching threshold set
Ct,x: cycle for target gene reaching threshold
Kx: a constant number
Equation 3: (for reference gene): Rt=R0(1+Er)Ct,r =Kr
Rt: number of reference gene reaching threshold
Ct,r: cycle for reference gene reaching threshold
Kx: another constant number
Xt over Rt is Xt/Rt= X0×(1+Ex)Ct,x / R0×(1+Er)Ct,r= Kx/ Kr=K
We suppose the amplification efficiency is the same for target and reference gene: Ex=Er=E, then
X0 / R0×(1+Er) Ct,x -Ct,r= K or ( Xn×(1+E)-ΔCt=K)
Xn: dealt initial number of target gene
ΔCt: difference in Ct for target and reference gene
We get: Xn=K×(1+E)-ΔCt
Xn(q) of each sample over reference sample Xn(cb):
Xn,q/Xn,cb=K×(1+E)-ΔCt,q/K×(1+E)ΔCt,cb=(1+E)-ΔΔCt
The advantage of this method is that standard curve is unnecessary but with two requirements:
1. Amplification efficiency for target and reference gene are the same.
2. Efficiency is the highest, nearly 100%.
To reach these, optimum operation is reached.
According to the results, we got:
-ΔΔCt(I20260)= 0.72773999
-ΔΔCt(J23115)= -2.169888735
Therefore, expression of GFP in I20260 is 20.72773999 time J23101 which is 1.6560, and for J23115 are 2-2.169888735 which is 0.2222.
We make a histogram about the fluorescence intensity of three samples below:
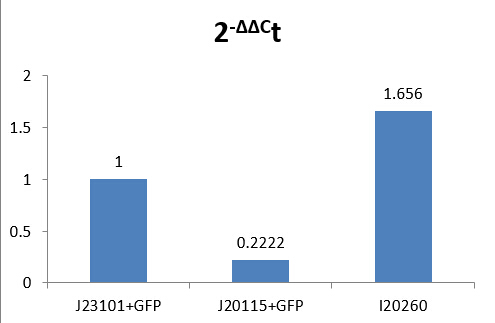
Fig 1.4 Comparison
Because there are inevitable differences in RNA extraction, and we cannot controlled all variables. According to a page on Part Registry and by consulting our advisors, we think those data of measurement as believable.
 "
"