Team:Aachen/Notebook/Software/Measurarty
From 2014.igem.org
(→Source Code) |
|||
| (25 intermediate revisions not shown) | |||
| Line 85: | Line 85: | ||
First, ''Measurarty'' segments the target image using '''Statistical Region Merging (SRM)''' in order to find regions of similar properties. After this step, we can segment the picture using '''histogram thresholding''' in [http://en.wikipedia.org/wiki/HSL_and_HSV HSV color space] to find candidate regions for pathogens. | First, ''Measurarty'' segments the target image using '''Statistical Region Merging (SRM)''' in order to find regions of similar properties. After this step, we can segment the picture using '''histogram thresholding''' in [http://en.wikipedia.org/wiki/HSL_and_HSV HSV color space] to find candidate regions for pathogens. | ||
Finally, a classification algorithm can detect the pathogen on our chips. | Finally, a classification algorithm can detect the pathogen on our chips. | ||
| + | |||
| + | To demonstrate the algorithm, the following sample image will be discussed. | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_test.jpg|title=Image taken from WatsOn to be analyzed by ''Measurarty'' algorithm|width=700px}} | ||
| Line 98: | Line 102: | ||
Compared to other clustering algorithms, SRM is quite leight weight, yet delivers ''deterministic'' results and is not dependent on a certain seed (like ''k''-means, for example). | Compared to other clustering algorithms, SRM is quite leight weight, yet delivers ''deterministic'' results and is not dependent on a certain seed (like ''k''-means, for example). | ||
| - | On the other hand, it can create as many refinements as one wants and is thus flexible enough for the our purposes. Finally there's already been knowledge about this algorithm in the group. | + | On the other hand, it can create as many refinements as one wants and is thus flexible enough for the our purposes. Finally, there's already been knowledge about this algorithm in the group. |
Statistical Region Merging (SRM) (Nook and Nielson, 2004) is a clustering technique also used directly for image segmentation. | Statistical Region Merging (SRM) (Nook and Nielson, 2004) is a clustering technique also used directly for image segmentation. | ||
| Line 128: | Line 132: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
| + | Finally, if applied to our test-image, regions are created and homogenoues regions form: | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_srmed.png|title=Regions created with SRM clustering|width=700px}} | ||
| Line 150: | Line 158: | ||
The remainder of this stage creates a mask of pixels that fulfill the conditions. | The remainder of this stage creates a mask of pixels that fulfill the conditions. | ||
| - | |||
| - | |||
<html> | <html> | ||
| Line 191: | Line 197: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | |||
| + | If you apply this HSV masking code to the SRMed test image, the following is created: | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_masked.png|title=Masked image|width=700px}} | ||
| Line 224: | Line 234: | ||
=== Empirical Evaluation === | === Empirical Evaluation === | ||
| - | Using our MATLAB code we found the lower threshold for the smoothness index to be $TS_l = 0.85$ and the upper threshold $TS_u = \infty$. | + | Using our MATLAB code, we found the lower threshold for the smoothness index to be $TS_l = 0.85$ and the upper threshold $TS_u = \infty$. |
Similarly, for $TI_l = 235$ and $TI_u = \infty$. | Similarly, for $TI_l = 235$ and $TI_u = \infty$. | ||
Using these settings, we can find a response already in images taken after 42 minutes. | Using these settings, we can find a response already in images taken after 42 minutes. | ||
| - | Ideally, one would rate the quality of the image segmentation using some ground truth, such as manual delineations. This still has to be | + | Ideally, one would rate the quality of the image segmentation using some ground truth, such as manual delineations. This still has to be implemented for our method. |
However, from visual observations, our method is showing promising results. | However, from visual observations, our method is showing promising results. | ||
| Line 280: | Line 290: | ||
</html> | </html> | ||
| + | This code actually creates two intermediate images from which the similarity index is calculated. | ||
| + | First the smoothed (disk-filter) input image is created and stored: | ||
| - | {{Team:Aachen/ | + | {{Team:Aachen/Figure|align=center|Aachen_meas_smoothed.png|title=Smoothed image|width=700px}} |
| + | |||
| + | Only white regions are candidate regions. | ||
| + | After smoothing, the similarity index is calculated. As expected, edges are detected and limit the area from which the target region can be selected. | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_smiliarity.png|title=Smoothness Index|width=700px}} | ||
| + | |||
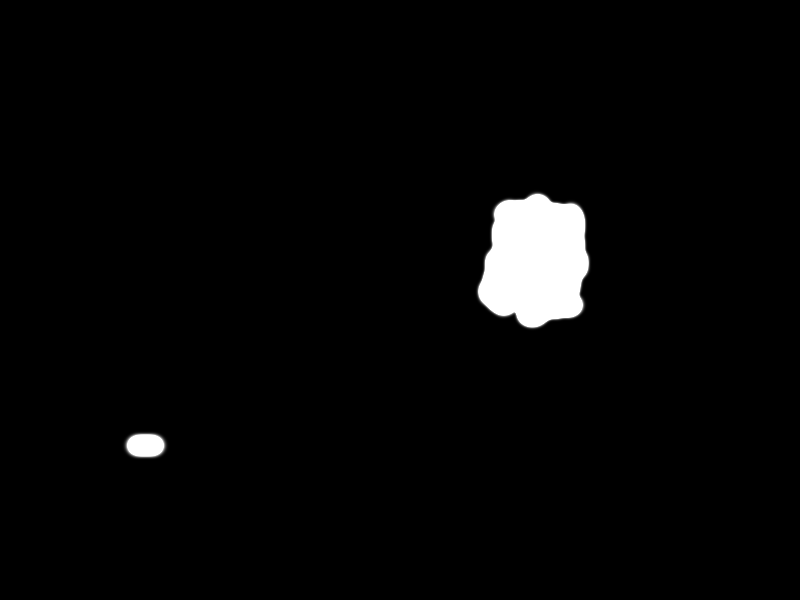
| + | Finally the selected pathogen region is selected by the black area in the following picture: | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_mask.png|title=Selected region|width=700px}} | ||
| + | |||
| + | Combined with the input image, the final segmentation is received: | ||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen_meas_final.png|title=Final the analyzed image|width=700px}} | ||
[[File:Aachen_14-10-15_Medal_Cellocks_iNB.png|right|150px]] | [[File:Aachen_14-10-15_Medal_Cellocks_iNB.png|right|150px]] | ||
| + | |||
| + | |||
| + | {{Team:Aachen/BlockSeparator}} | ||
== Achievements == | == Achievements == | ||
| Line 292: | Line 320: | ||
For example, changing from green to red fluorescence, only means to change the ''createMask'' function to select another target area. | For example, changing from green to red fluorescence, only means to change the ''createMask'' function to select another target area. | ||
| - | Overall the results | + | Overall the results are convincing. We have not yet performed a comparison to a manual delineation, however, by eye the results look promising and have a low error. |
Talking about computational complexity, the MATLAB code of course performs better than our own C++ implementation, which must be regarded as a proof-of-principle. | Talking about computational complexity, the MATLAB code of course performs better than our own C++ implementation, which must be regarded as a proof-of-principle. | ||
| Line 302: | Line 330: | ||
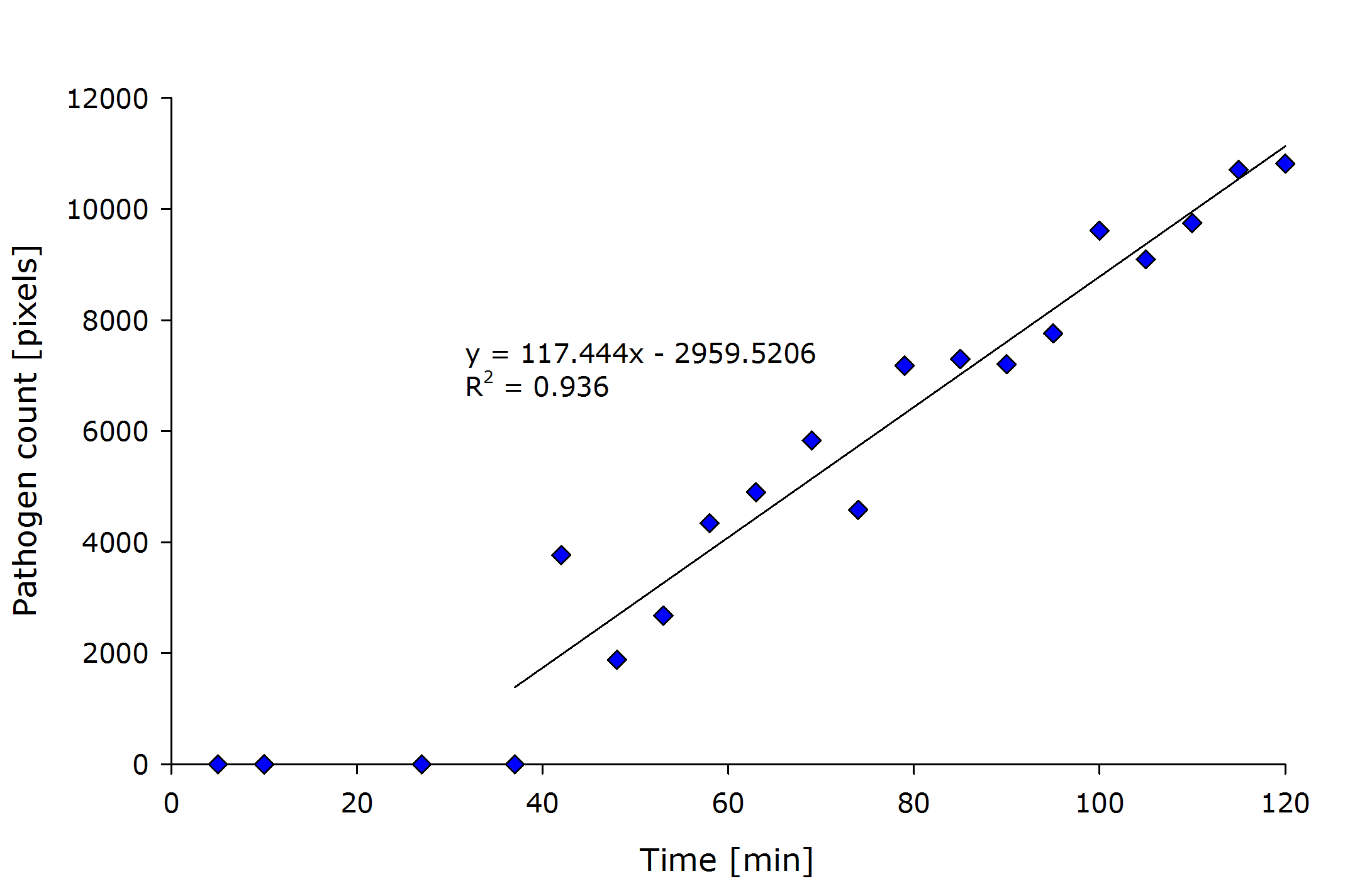
Due to the merging step in our C++-SRM algorithm implementation, our code has to do $O(x^2 \cdot y^2)$ comparisons, which then finally results in a runtime complexity of $O( x^2 \cdot y^2)$. | Due to the merging step in our C++-SRM algorithm implementation, our code has to do $O(x^2 \cdot y^2)$ comparisons, which then finally results in a runtime complexity of $O( x^2 \cdot y^2)$. | ||
| - | + | {{Team:Aachen/Figure|align=center|Aachen_meas_sizes.png|title=Pixel count of the detected pathogenic region versus time after induction.|width=700px}} | |
From the above figure it can also be seen that the detected amount of pathogenic-area correlates with time after induction. | From the above figure it can also be seen that the detected amount of pathogenic-area correlates with time after induction. | ||
The lag-phase can be explained first by the lag-phase of the cells, which first need to generate a response to the pathogen, and on the other hand, by too low fluorescence which is not detectable. | The lag-phase can be explained first by the lag-phase of the cells, which first need to generate a response to the pathogen, and on the other hand, by too low fluorescence which is not detectable. | ||
The pixel count also meets the expectation when looking at the sample files by eye. | The pixel count also meets the expectation when looking at the sample files by eye. | ||
| + | |||
| + | <center> | ||
| + | <div class="figure" style="float:{{{align|center}}}; margin: 0px 10px 10px 0px; border:{{{border|0px solid #aaa}}};width:{{{width|960px}}};padding:10px 10px 0px 0px;"> | ||
| + | {| | ||
| + | |<html> <img src="https://static.igem.org/mediawiki/2014/f/fc/Aachen_Measurarty_combined_slow.gif" width="960px"></html> | ||
| + | |- | ||
| + | |'''{{{title|Detecting ''P. aeroginosa'' with K131026}}}'''<br />{{{subtitle|The left half shows the original images from the device and the right half shows the same pictures with the detected pathogenic region analyzed by ''Measurarty''.}}} | ||
| + | |} | ||
| + | </div> | ||
| + | </center> | ||
It can be concluded that the ''Measurarty'' pipeline defines a robustly working chip-analysis algorithm which can detect pathogens from images supplied by ''WatsOn''. | It can be concluded that the ''Measurarty'' pipeline defines a robustly working chip-analysis algorithm which can detect pathogens from images supplied by ''WatsOn''. | ||
| Line 321: | Line 359: | ||
''Measuarty'' is the image analysis logic behind our project. It has been prototyped and developed in [http://www.mathworks.de/academia/student-competitions/igem/ MATLAB], and only later been ported into our ''WatsOn'' GUI. | ''Measuarty'' is the image analysis logic behind our project. It has been prototyped and developed in [http://www.mathworks.de/academia/student-competitions/igem/ MATLAB], and only later been ported into our ''WatsOn'' GUI. | ||
| - | We are happy to provide you with a zip-ped download of our MATLAB code here, as well as on the iGEM | + | We are happy to provide you with a zip-ped download of our MATLAB code here, as well as on the iGEM software repository on [https://github.com/orgs/igemsoftware/teams/aachen2014 github]. |
| - | * MATLAB code | + | * [https://static.igem.org/mediawiki/2014/6/6e/Aachen_measurarty.zip MATLAB code] |
| - | * link [https://github.com | + | * link [https://github.com/igemsoftware/AachenSoftProject2014/tree/master/measurarty github] |
| - | For the C++ conversion please see [https://2014.igem.org/Team:Aachen/Notebook/Engineering/WatsOn#watsonsoftware our WatsOn Software] section. | + | For the C++ conversion please see [https://2014.igem.org/Team:Aachen/Notebook/Engineering/WatsOn#watsonsoftware our ''WatsOn'' Software] section. |
=== Using the MATLAB Code === | === Using the MATLAB Code === | ||
Latest revision as of 03:46, 18 October 2014
|
|
|
|
|
|
|
 "
"