Team:Aachen/Collaborations/Heidelberg
From 2014.igem.org
m (→The Team Heidelberg expression vectors) |
(→Attempting to Characterize the pSBX vectors) |
||
| (5 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Team:Aachen/Header}} | {{Team:Aachen/Header}} | ||
| - | = | + | = Testing [[Team:Heidelberg|Team Heidelberg]]s pSBX-vectors = |
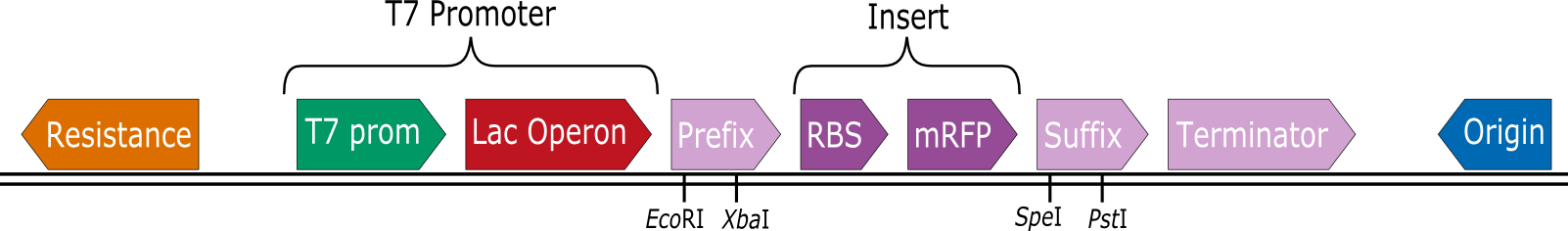
The iGEM team Heidelberg constructed a set of expression vectors that contain a T7 promoter prior to the BioBrick prefix. This way RBS-CDS parts can be cloned into the vectors by standard BioBrick assembly and then expressed in BL21(DE3) by induction with IPTG. | The iGEM team Heidelberg constructed a set of expression vectors that contain a T7 promoter prior to the BioBrick prefix. This way RBS-CDS parts can be cloned into the vectors by standard BioBrick assembly and then expressed in BL21(DE3) by induction with IPTG. | ||
| Line 10: | Line 10: | ||
In collaboration with the iGEM Team Heidelberg the iGEM Team Aachen tested 8 of these vectors for their expression capabilities. | In collaboration with the iGEM Team Heidelberg the iGEM Team Aachen tested 8 of these vectors for their expression capabilities. | ||
| - | == | + | We successfully cloned and expressed one of our fusion proteins K139020 in a pSBX1A3 vector. You can read more about the background of this expression on the [[Team:Aachen/Project/Gal3|Galectin-3 page]]. |
| + | |||
| + | |||
| + | == Attempting to Characterize the pSBX vectors == | ||
These are the tested constructs: | These are the tested constructs: | ||
| Line 38: | Line 41: | ||
To find out more about the Results of the characterization check out the the [https://2014.igem.org/Team:Heidelberg/Team/Collaborations collaboration page of Team Heidelberg]. | To find out more about the Results of the characterization check out the the [https://2014.igem.org/Team:Heidelberg/Team/Collaborations collaboration page of Team Heidelberg]. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="figure" style="float:center; margin: 10px 10px 10px 40px; border:0px solid #aaa;width:600px;padding:0px;"> | <div class="figure" style="float:center; margin: 10px 10px 10px 40px; border:0px solid #aaa;width:600px;padding:0px;"> | ||
| Line 59: | Line 46: | ||
|{{Team:Aachen/Test/GalleryHeader}} | |{{Team:Aachen/Test/GalleryHeader}} | ||
<html> | <html> | ||
| - | <div id="slider1_container" style="position: relative; width: 600px; height: 500px; background-color: # | + | <div id="slider1_container" style="position: relative; width: 600px; height: 500px; background-color: #FFF; overflow: hidden; "> |
| Line 75: | Line 62: | ||
overflow: hidden;"> | overflow: hidden;"> | ||
<div> | <div> | ||
| - | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/ | + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/f/f3/Aachen_HDSummary.PNG"/></a> |
</div> | </div> | ||
<div> | <div> | ||
| - | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/ | + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/a/a5/Aachen_PSBX1A3.PNG"/></a> |
</div> | </div> | ||
<div> | <div> | ||
| - | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/ | + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/3/34/Aachen_PSBX1C3.PNG"/></a> |
</div> | </div> | ||
<div> | <div> | ||
| - | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/ | + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/1/13/Aachen_PSBX4A5.PNG"/></a> |
</div> | </div> | ||
<div> | <div> | ||
| - | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/ | + | <a u=image href="#"><img src="https://static.igem.org/mediawiki/2014/2/22/Aachen_PSBX4C5.PNG"/></a> |
</div> | </div> | ||
</div> | </div> | ||
| Line 102: | Line 89: | ||
</html> | </html> | ||
|- | |- | ||
| - | |'''Fluorescence data of pSBX cultivation'''<br /> | + | |'''Fluorescence data of pSBX cultivation'''<br />While the positive control did show an increase of fluorescence to a maximum of ca. 20000 relative fluorescence units, there appeared to be something wrong with the pSBX expression strains. We did not have the time to check what exactly was wrong with the cells, but it's not because of the vectors, because we successfully used the pSBX1A3 to express our K1319020 fusion protein. |
|} | |} | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Latest revision as of 21:51, 17 October 2014
|
 "
"