Team:Oxford/Parts
From 2014.igem.org
(Difference between revisions)
| (21 intermediate revisions not shown) | |||
| Line 21: | Line 21: | ||
</div> | </div> | ||
<div style="background-color:white; border-bottom-left-radius:10px;border-bottom-right-radius:10px; padding-left:10px;padding-right:10px;min-width:300px;margin-top:100px;"> | <div style="background-color:white; border-bottom-left-radius:10px;border-bottom-right-radius:10px; padding-left:10px;padding-right:10px;min-width:300px;margin-top:100px;"> | ||
| - | + | <br> | |
| - | Oxford iGEM has submitted three parts to the registry, which | + | <h1>Oxford iGEM has submitted three parts to the registry, which are described on the relevant registry pages. </h1> |
<br><br> | <br><br> | ||
<a href="http://parts.igem.org/Part:BBa_K1446001"> | <a href="http://parts.igem.org/Part:BBa_K1446001"> | ||
| - | <u>Part no. 1(BBa_K1446001)</u></a> is the gene coding for DcmA with an N-terminal pdu-microcompartment tag. | + | <u>Part no. 1 (BBa_K1446001)</u></a> is the gene coding for DcmA with an N-terminal pdu-microcompartment tag. |
| + | <img src="https://static.igem.org/mediawiki/parts/c/c0/BBa_k1446001.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
<br><br> | <br><br> | ||
<a href="http://parts.igem.org/Part:BBa_K1446002"> | <a href="http://parts.igem.org/Part:BBa_K1446002"> | ||
| - | <u>Part no. 2(BBa_K1446002)</u></a> is a superfolder GFP-gene with the N-terminal pdu-microcompartment tag. We have included a ribosome binding site upstream of the coding region | + | <u>Part no. 2 (BBa_K1446002)</u></a> is a superfolder GFP-gene with the N-terminal pdu-microcompartment tag. We have included a ribosome binding site upstream of the coding region as well as a his-tag at the C-terminus. This part has been used for fluorescent image analysis to show that the targeting mechanism of the N-terminal microcompartment tag works as expected. |
| + | <img src="https://static.igem.org/mediawiki/parts/1/1d/BBa_k1446002.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
<br><br> | <br><br> | ||
<a href="http://parts.igem.org/Part:BBa_K1446003"> | <a href="http://parts.igem.org/Part:BBa_K1446003"> | ||
| - | <u>Part no. 3(BBa_K1446003)</u></a> is the dcmR gene with a tetR promoter. We have used this part to get fluorescence data for our model described here. | + | <u>Part no. 3 (BBa_K1446003)</u></a> is the dcmR gene with a tetR promoter. We have used this part to get fluorescence data for our model described <a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show2"> |
| + | <u>here</u></a> and showed that mCherry expression increased at higher ATC (anhydrous tetracycline) concentration, as expected. | ||
| + | <img src="https://static.igem.org/mediawiki/parts/8/80/BBa_k1446003.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
| - | <br><br | + | <br><br> |
Parts no. 1 and 2 belong to our bioremediation subproject and part 3 to our biosensor subproject. | Parts no. 1 and 2 belong to our bioremediation subproject and part 3 to our biosensor subproject. | ||
<br> | <br> | ||
<br> | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <h1>Oxford iGEM have also constructed further parts but were not fully in a BioBrick compatible plasmid thus were not sent before the deadline but will be on the registry in the near future</h1> | ||
| + | <br> | ||
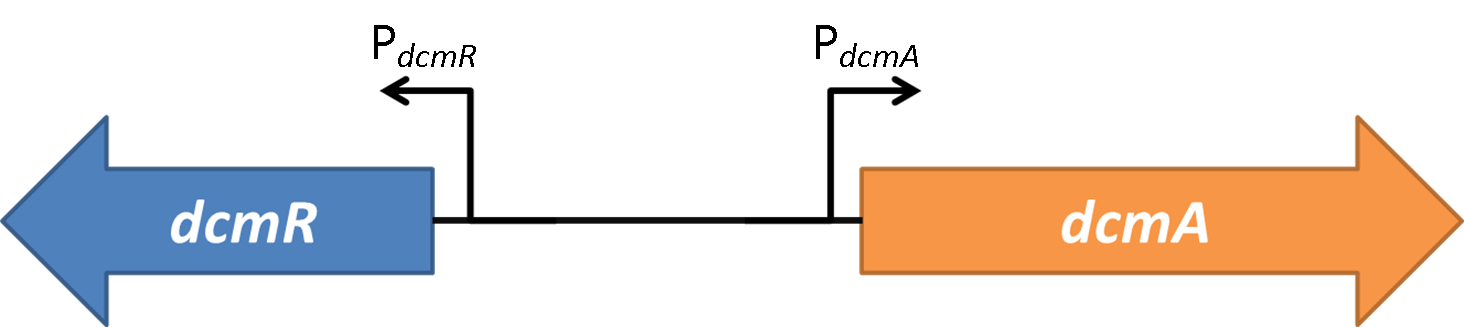
| + | The following two constructs are based on the intergenic region shown below from the native DCM-degrading bacterium (Methylobacterium extorquens DM4): | ||
| + | <img src="https://static.igem.org/mediawiki/2014/7/7f/Oxford_charac3.png" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;" /> | ||
| + | |||
| + | <h1>PdcmA-sfGFP</h1> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/e/e8/Oxfordigem_pdcma.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
| + | <br> | ||
| + | This construct involves the bidirectional promoter that has been characterised as a new tool for molecular biologists. In this construct sfGFP is placed under control in the PdcmA direction. | ||
| + | Click <a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show11"> | ||
| + | <u>here</u></a> to see characterisation on this part in plasmid PJ404. | ||
| + | <br><br> | ||
| + | <br> | ||
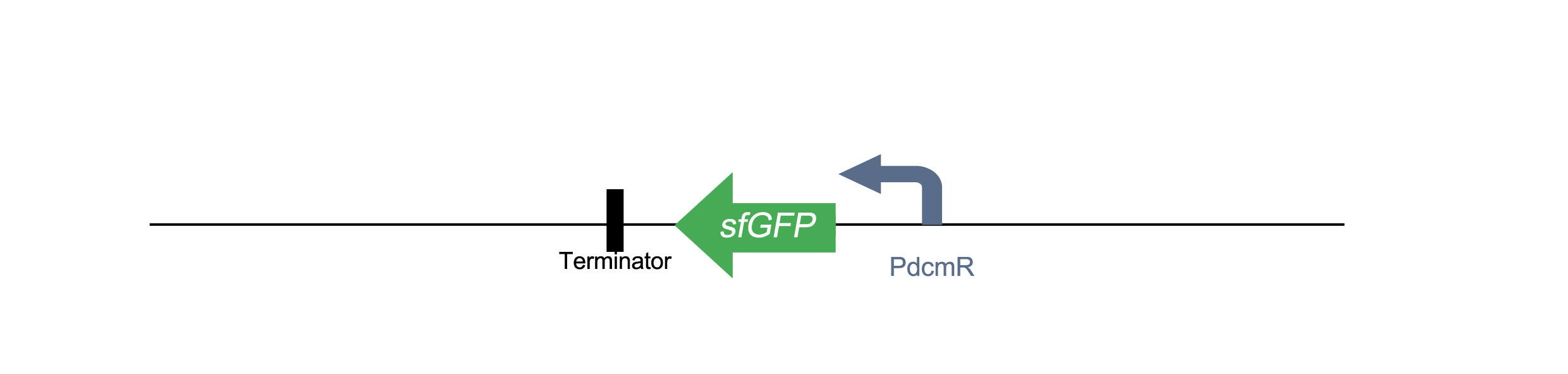
| + | <h1>PdcmR-sfGFP</h1> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/ba/Oxfordigem_pdcmR.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
| + | <br> | ||
| + | Similar to the construct above, this involves the same bidirectional promoter with sfGFP in placed in the opposite direction under control of PdcmR. | ||
| + | Click <a href="https://2014.igem.org/Team:Oxford/biosensor_characterisation#show11"> | ||
| + | <u>here</u></a> to see characterisation on this part in plasmid PJ404. | ||
| + | <br> | ||
| + | <br> | ||
| + | <h1>dcmA-sfGFP</h1> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/9/93/DcmA-sfGFP.jpg" style="float:left;position:relative; width:90%; margin-right:75%;margin-bottom:2%;" /> | ||
| + | <br> | ||
| + | For the <a href="https://2014.igem.org/Team:Oxford/codon_optimisation#show14"> | ||
| + | <u>analysis</u></a> of dcmA-expression we have used dcmA-fused to sfGFP in the pRSFDuet vector, which has a T7-promoter that is under lac-control. | ||
| + | <br><br> | ||
| + | |||
| + | |||
</div> | </div> | ||
<br> | <br> | ||
Latest revision as of 03:47, 18 October 2014
 "
"