Team:Freiburg/Content/Results/The combination
From 2014.igem.org
Mirja Harms (Talk | contribs) |
|||
| (20 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
<section> | <section> | ||
| - | + | <p> | |
| - | + | Our ultimate goal is to combine the spatial resolution of the light system with the specificity of our viral vector to generate patterns in a homogeneous cell culture. Since cells that are not illuminated with the inducing light should not express the mCAT-1 receptor, they should not get infected by viral particles. | |
| + | </p> | ||
| - | <h2 id="Light induced receptor">Light | + | <h2 id="Light-induced-receptor">Light Induced Receptor</h2> |
| - | <p> | + | <p>For light induced gene transfer with our viral vector we cloned mCAT-1, the receptor our viral vector is specific for, into the response plasmid for the blue light system (plasmid p14ls_003 coding for mCAT-1-HA-P2A-mCherry under the control of the Gal4 upstream activating sequence and a minimal CMV promoter) or the red light system (p14rz_002 coding for mCAT-1-HA-P2A-mCherry under the control of the tet operon and a minimal CMV promoter). These P2A sequence between the mCAT-1-HA receptor and the mCherry reporter induces a co-translational self-cleavage, leaving behind the receptor-HA tag fusion and the cytosolic mCherry tag. The activation of gene expression was induced by illumination with 660 nm for the red light system (Fig. 1) and 465 nm for the blue light system (Figs. 2, 3). When the cells containing the light inducible receptor were not exposed to light (dark controls), no mCherry was detected in the cytosol, i.e. no receptor was expressed and transported to the cell surface. |
</p> | </p> | ||
| Line 23: | Line 24: | ||
</a> | </a> | ||
<figcaption> | <figcaption> | ||
| - | <p class="header">Red light induced receptor.</p> | + | <p class="header">Figure 1: Red light induced receptor in CHO cells.</p> |
| - | <p class="desc">CHO cells were transfected with the red light system (PKM022) and the light induced receptor (p14rz_002). The receptor was labeled with mCherry; (left) after illumination with red light, (middle) | + | <p class="desc">CHO cells were transfected with the red light system (PKM022) and the light induced receptor (p14rz_002). The receptor was labeled with mCherry; (left) after illumination with red light, (middle) negative control left in the dark, (right) positive control. Cells were stained with DAPI. <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Confocal_Red_Light">Labjournal</a></p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | |||
| + | |||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | |||
| + | <p>Since our viral vector was not able to integrate into cells not expressing mCAT-1 on their surface, it was not able to infect non-illuminated cells (Figs. 2, 3). As a reporter to detect the cells that were infected by viral particles, we used EGFP as a cargo. In principle any gene of interest could be transduced under light control using The AcCELLerator. | ||
| + | </p> | ||
<figure class="fig-full-width"> | <figure class="fig-full-width"> | ||
| Line 33: | Line 41: | ||
</a> | </a> | ||
<figcaption> | <figcaption> | ||
| - | <p class="header">Blue light induced receptor.</p> | + | <p class="header">Figure 2: Blue light induced receptor in HEK-293T.</p> |
| - | <p class="desc">HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry. Cells were infected with MuLV EGFP afterwards; (left) incubation in the dark, (middle) after illumination with blue light, (right) cells expressing the light induced receptor were infected with MuLV. <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Controls_Blue_Light">Labjournal</a></p> | + | <p class="desc">HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry. Cells were infected with MuLV-EGFP afterwards; (left) incubation in the dark, (middle) after illumination with blue light, (right) cells expressing the light induced receptor were infected with MuLV. <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Controls_Blue_Light">Labjournal</a></p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div class="col-sm-6"> | ||
| + | <figure class="fig-full-width"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/4/4b/Freiburg2014_Receptor_10_light_induced_plus_Virus_singel_channels2.png"> <!-- ORGINAL --> | ||
| + | <img class="img-no-pad" src="https://static.igem.org/mediawiki/2014/4/4b/Freiburg2014_Receptor_10_light_induced_plus_Virus_singel_channels2.png"> <!-- Thumbnail --> | ||
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header">Figure 3: Blue light induced receptor in HEK-293T</p> | ||
| + | <p class="desc">HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry that was cleaved during expression, thus remaining in the cytoplasm. Cells were infected with MuLV EGFP afterwards. (A) Overlay of all three channels, (B) DAPI stained nuclei, (C) EGFP expression in infected cells, (D) mCherry expression. Objective plan apo 60x, NA 1.40. <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Confocal_virus">Labjournal</a></p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| - | <p> | + | <div class="row category-row"> |
| + | <div class="col-sm-6"> | ||
| + | <p>We determined the expression time of the receptor that was induced by blue light after illumination for five hours. We used a receptor that was labeled with a fused HA-tag and mCherry separated by a P2A site (p14ls_003), for analysis with Western blot and fluorescence microscopy. The receptor had an expression peak at 24 hours after beginning of illumination. | ||
</p> | </p> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
<figure> | <figure> | ||
<a href="https://static.igem.org/mediawiki/2014/b/bb/Freiburg2014-10-14_bluel-kinetic-microscopy.jpg"> <!-- ORGINAL --> | <a href="https://static.igem.org/mediawiki/2014/b/bb/Freiburg2014-10-14_bluel-kinetic-microscopy.jpg"> <!-- ORGINAL --> | ||
| Line 46: | Line 73: | ||
</a> | </a> | ||
<figcaption> | <figcaption> | ||
| - | <p class="header"> | + | <p class="header">Figure 4: Kinetics of the blue light induced receptor.</p> |
| - | <p class="desc">HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003, mCherry | + | <p class="desc">HEK-293T cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003, mCherry linked receptor). Pictures were taken after 12h, 15h, 18h and 24h. <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Blue_Light_Kinetik">Labjournal</a></p> |
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <h2 id="Results-LightSystem-MuLVSEAP">Pattern Generation</h2> | ||
| + | |||
| + | <p>For generating pattern in homogeneous cell layers, we transfected HEK293T cells with the blue light system and the blue light induced receptor (p14ls_003). Dishes were covered with a photo mask preventing areas in the cell culture from light exposure. However, the blue light system is very sensitive to even low intensities of blue light. Due to scatterd light, the receptor was activated in a large area of the cell culture. Thus patterns were not visible at this time point due to activation of receptor expression by scattered light (Fig. 5). Therefore, we also did not apply the virus to these cultures. | ||
| + | </p> | ||
| + | |||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/3/3c/Freiburg2014-09-30_HEK_30min_BF_stripe_bearbeitet.jpg"> <!-- ORGINAL --> | ||
| + | <img class="img-no-pad" src="https://static.igem.org/mediawiki/2014/3/3c/Freiburg2014-09-30_HEK_30min_BF_stripe_bearbeitet.jpg"> <!-- Thumbnail --> | ||
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header">Figure 5: The light induced receptor was induced by light scatter.</p> | ||
| + | <p class="desc">HEK293T cells were transfected with the light system and the light induced receptor (p14ls_003) labeled with mCherry. Cells were illuminated using a photo mask that covered parts of the cell culture and prevented them from light exposure. However, the receptor expression was activated also outside of the pattern.<a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Light_scattered_acitivated_receptor">Labjournal</a></p> | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| - | |||
</section> | </section> | ||
Latest revision as of 03:35, 18 October 2014
The Combination
Our ultimate goal is to combine the spatial resolution of the light system with the specificity of our viral vector to generate patterns in a homogeneous cell culture. Since cells that are not illuminated with the inducing light should not express the mCAT-1 receptor, they should not get infected by viral particles.
Light Induced Receptor
For light induced gene transfer with our viral vector we cloned mCAT-1, the receptor our viral vector is specific for, into the response plasmid for the blue light system (plasmid p14ls_003 coding for mCAT-1-HA-P2A-mCherry under the control of the Gal4 upstream activating sequence and a minimal CMV promoter) or the red light system (p14rz_002 coding for mCAT-1-HA-P2A-mCherry under the control of the tet operon and a minimal CMV promoter). These P2A sequence between the mCAT-1-HA receptor and the mCherry reporter induces a co-translational self-cleavage, leaving behind the receptor-HA tag fusion and the cytosolic mCherry tag. The activation of gene expression was induced by illumination with 660 nm for the red light system (Fig. 1) and 465 nm for the blue light system (Figs. 2, 3). When the cells containing the light inducible receptor were not exposed to light (dark controls), no mCherry was detected in the cytosol, i.e. no receptor was expressed and transported to the cell surface.
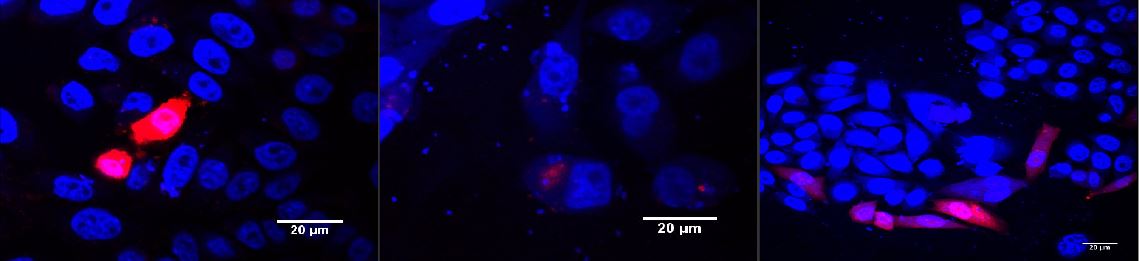
Figure 1: Red light induced receptor in CHO cells.
CHO cells were transfected with the red light system (PKM022) and the light induced receptor (p14rz_002). The receptor was labeled with mCherry; (left) after illumination with red light, (middle) negative control left in the dark, (right) positive control. Cells were stained with DAPI. Labjournal
Since our viral vector was not able to integrate into cells not expressing mCAT-1 on their surface, it was not able to infect non-illuminated cells (Figs. 2, 3). As a reporter to detect the cells that were infected by viral particles, we used EGFP as a cargo. In principle any gene of interest could be transduced under light control using The AcCELLerator.

Figure 2: Blue light induced receptor in HEK-293T.
HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry. Cells were infected with MuLV-EGFP afterwards; (left) incubation in the dark, (middle) after illumination with blue light, (right) cells expressing the light induced receptor were infected with MuLV. Labjournal

Figure 3: Blue light induced receptor in HEK-293T
HEK cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003). The receptor was labeled with mCherry that was cleaved during expression, thus remaining in the cytoplasm. Cells were infected with MuLV EGFP afterwards. (A) Overlay of all three channels, (B) DAPI stained nuclei, (C) EGFP expression in infected cells, (D) mCherry expression. Objective plan apo 60x, NA 1.40. Labjournal
We determined the expression time of the receptor that was induced by blue light after illumination for five hours. We used a receptor that was labeled with a fused HA-tag and mCherry separated by a P2A site (p14ls_003), for analysis with Western blot and fluorescence microscopy. The receptor had an expression peak at 24 hours after beginning of illumination.

Figure 4: Kinetics of the blue light induced receptor.
HEK-293T cells were transfected with the blue light system (PKM292 and PKM297) and the light induced receptor (p14ls_003, mCherry linked receptor). Pictures were taken after 12h, 15h, 18h and 24h. Labjournal
Pattern Generation
For generating pattern in homogeneous cell layers, we transfected HEK293T cells with the blue light system and the blue light induced receptor (p14ls_003). Dishes were covered with a photo mask preventing areas in the cell culture from light exposure. However, the blue light system is very sensitive to even low intensities of blue light. Due to scatterd light, the receptor was activated in a large area of the cell culture. Thus patterns were not visible at this time point due to activation of receptor expression by scattered light (Fig. 5). Therefore, we also did not apply the virus to these cultures.

Figure 5: The light induced receptor was induced by light scatter.
HEK293T cells were transfected with the light system and the light induced receptor (p14ls_003) labeled with mCherry. Cells were illuminated using a photo mask that covered parts of the cell culture and prevented them from light exposure. However, the receptor expression was activated also outside of the pattern.Labjournal
 "
"