|
|
| (68 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | <html> | | <html> |
| | <div id="project-biobrickbox-teaser" class="full-width teaser-img"> </div> | | <div id="project-biobrickbox-teaser" class="full-width teaser-img"> </div> |
| | + | <section> |
| | + | <section> |
| | + | <div> |
| | + | </html> |
| | + | |
| | + | = ''Bacillus'' BioBrickBox = |
| | + | In 2012, the iGEM-team LMU-Munich began the task to develop essential BioBricks, like vectors, promoters, reporters and affinity tags, especially suited for the use in ''Bacillus subtilis'' in order to establish a new chassis in the ''Escherichia coli''-dominated world of iGEM. Even after the finals of the iGEM competition in 2012, the project was pursued further since its importance for synthetic biology and resulted in a publication in the Journal of Biological Engineering about this so called [http://www.jbioleng.org/content/7/1/29 ''Bacillus'' BioBrick Box]. |
| | + | |
| | + | This year, our team aims to enhance the ''Bacillus'' BioBrick Box by adding new parts, like different colored fluorescent proteins, a RBS-collection, evaluating protein linkers for ''B. subtilis'' and creating a redesigned backbone for resistance free strain generation. |
| | + | |
| | + | <html> |
| | + | </div> |
| | + | </section> |
| | + | <section class="bg-color-2"> |
| | + | <div> |
| | + | </html> |
| | + | |
| | + | == Resistance free strain generation == |
| | + | With BaKillus as an medical application it is most likely that it will be released - though in small amounts - into the environment. |
| | + | Therefore we sought after a solution in order to keep the final BaKillus free of unnecessary antibiotic resistances. In order to do so we developed a resistance free gene insertion strategy for integrative ''Bacillus subtilis'' vectors. |
| | + | |
| | + | <html> |
| | + | <section class="accordion"> |
| | + | <div> |
| | + | <input id="rfc-bground" name="rfc-accord" type="checkbox" /> |
| | + | <label for="rfc-bground">Background</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | The ''upp'' gene from Bacillus subtilis W168 encodes for a Uracilphosphoribosyl transferase (UPRTase). |
| | + | Its key reaction in uracil salvage is the reaction of a uracil molecule with a 5'-phosphoribosyl-a-1- pyrophosphate (PRPP) molecule, resulting in the formation of UMP. |
| | + | A second locus, the ''pyrR'' gene, encoding a second UPRTase has been identified. However it has been shown, that the UPRTase derived from ''pyrR'' locus has an influence on overall UPRTase activity of < 1 %. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC176586/] |
| | + | This makes the ''upp''-derived UPRTase the only physiologically relevant catalyst for UPRTase activity. |
| | + | Exposure to the pyrimidine analogue 5-Fluorouracil UPRTase results in production of 5-fluoro-dUMP, a very potent inhibitor of the thymidylate synthase (Neuhard, J. (1983) Utilization of preformed pyrimidine bases and nucleosides. In Metabolism of Nucleotides, Nucleosides and Nucleobases in Microorganisms. Munch- Petersen, A. (eds). New York: Academic Press, pp. 95– 148. ). As a result 5-FU is toxic to the ''Bacillus subtilis'' W168 strain. |
| | + | ''B. subtilis'' 5FU-resistant (5FUR) mutants selected on low drug concentration (10 mM 5FU) are UPRTase-defective. [http://www.nature.com/nature/journal/v390/n6657/abs/390249a0.html]. |
| | + | This has made ''upp'' a go to choice for negative selection in combination with an ''B. subtilis'' W168 ''Δupp'' strain. So far it has been used to make clean in-frame deletions and point mutations. [http://www.ncbi.nlm.nih.gov/pubmed/12366828] |
| | + | |
| | + | However, to our knowledge, no application using ''upp'' for clean insertions has been established so far. |
| | + | |
| | + | The I-SceI Restricition endonuclease has a highly specific recognition sequence of 18 nucleotides. No such sequence is present in the ''B.subtilis'' W168 strain. It creates a double strand break at targeted location, which leads to an increased rate of repair at the specific site. By this, the rate of homologous recombination is increased by a factor of 100. [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0081370] |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="rfc-design" name="rfc-accord" type="checkbox" /> |
| | + | <label for="rfc-design">Design</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | The Plan was to restructure the BioBrick compatible, integrative ''Bacillus subtilis'' vectors [http://parts.igem.org/Part:BBa_J179000 pSB1C], [http://parts.igem.org/Part:BBa_J179001 pSB2E] and [http://parts.igem.org/Part:BBa_J179002 pSB4S] in a fashion that leads to deletion of the antibiotic resistance after an insertion of a gene of interest has been generated. |
| | + | Integration of those basic vectors is achieved via homologous recombination between the ''amyE''/''lacA''/''thrC'' locus, respectively, and the corresponding up and down fragments on the vector. This leads to an insertion of the gene of interest within the RCF10 compatible multiple cloning site and the resistance for positive selection (Cm<sup>r</sup>/MLS<sup>r</sup>/Spec<sup>r</sup>). |
| | + | |
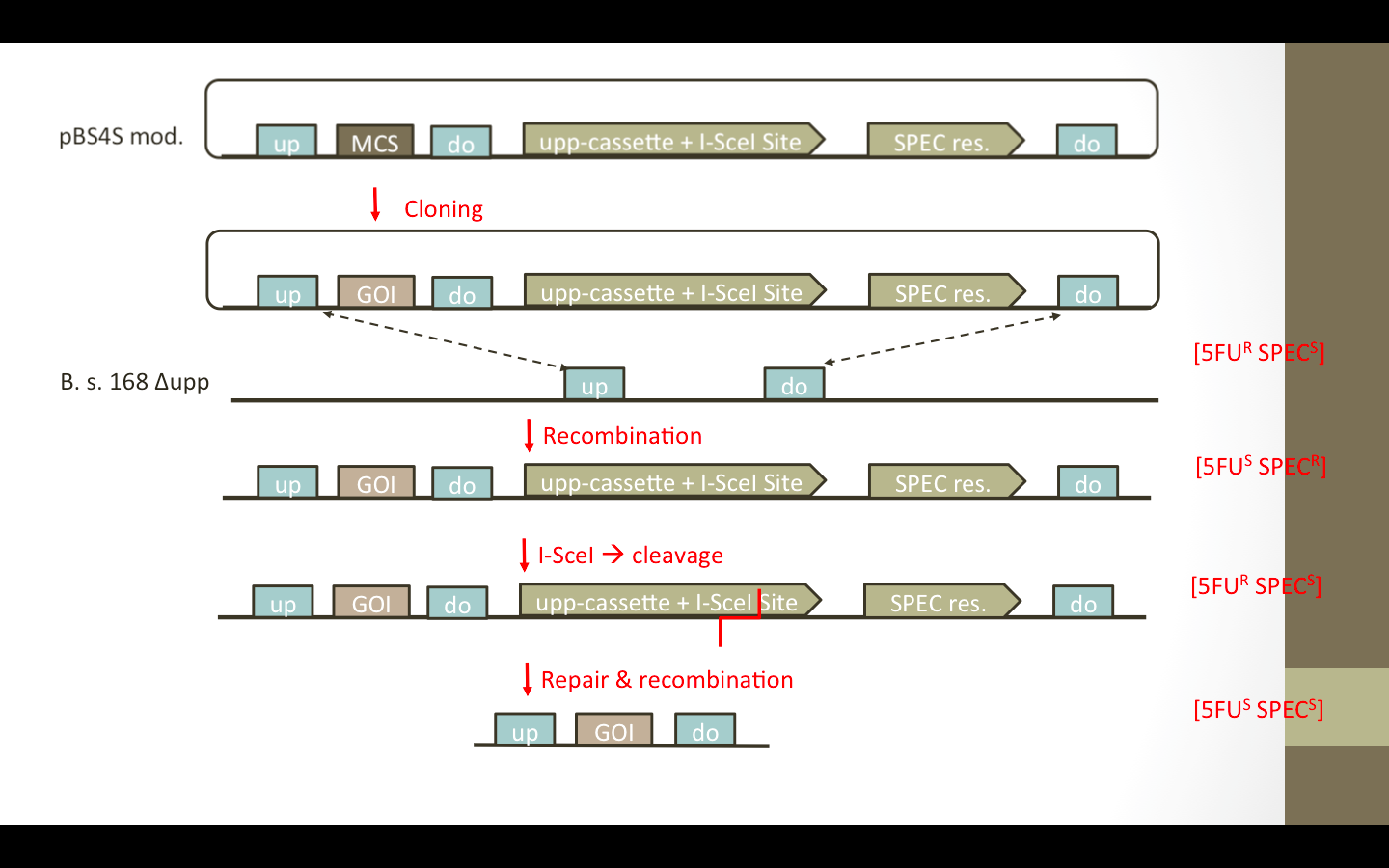
| | + | We wanted to add the ''upp''-cassette, containing an I-SceI site, as well as an additional up/down fragment to the vector. The desired vectors are presented in Fig. 1. The ''upp''-cassette will allow negative selection in 5FU media. The contained I-SceI site will be cut by the I-SceI restriction endonuclease which is encoded on a helper plasmid pEBS-cop1. [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0081370] (Figure) |
| | + | |
| | + | |
| | + | |
| | + | [[File:LMU14 RFCloning pEBS-cop1.jpg|thumb|center|pEBS-cop1. Plasmid containing the ''i-sceI'' gene]] |
| | + | Cloning Strategy |
| | + | |
| | + | [[File:LMU2014 BioBrickBox ResistanceFree Cloning Strategy.png|920px|center]] |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="rfc-results" name="rfc-accord" type="checkbox" /> |
| | + | <label for="rfc-results">Results</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | Lab work on this project only started at end of July |
| | + | So far the ''upp'' cassete has been modified to fullfill the Freiburg Standard (RCF25) criteria and was successfully sequencend and submitted to the registry as BioBrick part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1351023 BBa_K1351023]. |
| | + | Creation of the complete and functional backbones has not been successfull. We assume that due to the duplication of homologous sequences for the integration into ''Bacillus subtilis'' genomic loci a deletion has occured during phusion PCR. This is supported by the results of multiple digests of the resulting plasmid by different restriction enzymes. |
| | + | |
| | + | We plan on continuing work on this project after the iGEM Competition |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | </section> |
| | + | </div> |
| | + | </section> |
| | + | <section class="bg-color-3"> |
| | + | <div> |
| | + | </html> |
| | + | |
| | + | == Fluorescent Proteins == |
| | + | Fluorescent proteins (FPs) are important tools in research on a cellular or molecular basis and in the past years, many different FPs with very diverse qualities related to color, brightness, duration, bleaching, maturation time and other parameters were developed. |
| | + | Unfortunately, most of these FPs are optimized for use in ''E. coli'', yeast or mammalian cells, and we aim to evaluate the properties of different FPs for the use in ''B. subtilis'' to enhance our Bacillus BioBrick Box even further. |
| | + | <html> |
| | + | <section class="accordion"> |
| | + | <div> |
| | + | <input id="fps-bground" name="fps-accord" type="checkbox" /> |
| | + | <label for="fps-bground">Background</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | Fluorescent Proteins are often derivates of the first described FP, GFP, which was isolated from the jellyfish ''Aequorea victoria''. This protein is of typical barrel-shape, built from 11 ß-sheets and a chromophore in the middle of the barrel. This barrel structure is typical for fluorescent proteins, whether they derive from GFP or not. Other colors than green were developed by changing single amino acids in the chromophore of GFP (e.g.: Yellow: YFP and Cyan: CFP) or by analyzing fluorescent proteins from other organisms, like ''Discosoma striata'', a coral, from which some of our red FPs, like dsRed derive. |
| | + | FPs are an important tool in today’s research, for example in order to study gene expression or the location of specific proteins in a cell or a whole organism. This is the reason, why we tried to establish some different colors of FPs for the usilization in ''B. subtilis'' by evaluating them for the ''Bacillus'' BioBrick Box. |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="fps-design" name="fps-accord" type="checkbox" /> |
| | + | <label for="fps-design">Design</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | Seven different FPs (Table 1) where chosen and either obtained from the registry or from the lab group Mascher. |
| | + | The BioBrick <partinfo>BBa_E1010</partinfo>was mutated via site directed mutagenesis by overlap extension PCR in order to delete two AgeI-Restriction sites and make the BioBrick compatible for the Freiburg standard RFC25. This improved BioBrick is called <html><a href="http://parts.igem.org/Part:BBa_K1351021">BBa_K1351021</a></html> |
| | + | The BioBricks and [http://parts.igem.org/Part:BBa_K592100 BBa_K592100] (mTagBFP) where both provided with the necessary overhangs for the Freiburg standard, the other FPs where already provided with the proper restriction overhangs. |
| | + | These seven FPs where combined C- and N-terminally with a His-Tag ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K823037 BBa_K823037]) and cloned into the vector pBS0K-Pspac ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1351040 BBa_K1351040]) and transformed into ''B. subtilis''. |
| | + | |
| | + | |
| | + | |
| | + | {|cellspacing="0" border="1" |
| | + | |+ '''Table 1''': Used fluorescent proteins |
| | + | !BioBrick number |
| | + | !Name |
| | + | !Color |
| | + | !Excitation Peak |
| | + | !Emission Peak |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K1351021 BBa_K1351021] |
| | + | | DsRed |
| | + | |Red |
| | + | | align=right | 584nm |
| | + | | align=right | 607nm [1] |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K592100 BBa_K592100] |
| | + | | mTagBFP |
| | + | | Blue |
| | + | | align=right | 399nm |
| | + | | align=right | 456nm |
| | + | |- |
| | + | | [http://parts.igem.org/Part:BBa_K1159302 BBa_K1159302] |
| | + | | eCFP |
| | + | | Cyan |
| | + | | align=right | 439nm |
| | + | | align=right | 476nm |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K1159301 BBa_K1159301] |
| | + | | sYFP2 |
| | + | | Yellow |
| | + | | align=right | 515nm |
| | + | | align=right | 527nm |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K823039 BBa_K823039] |
| | + | | gfpmut1 |
| | + | |Green |
| | + | | align=right | 395nm |
| | + | | align=right | 509nm |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K1351042 BBa_K1351042] |
| | + | | mcherry |
| | + | | Red |
| | + | | align=right | 587nm |
| | + | | align=right | 610nm |
| | + | |- |
| | + | |[http://parts.igem.org/Part:BBa_K823029 BBa_K823029] |
| | + | | mKate 2 |
| | + | |Red |
| | + | | align=right | 588nm |
| | + | | align=right | 633nm |
| | + | |- |
| | + | |} |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | |
| | + | [[File:LMU14_FP_Gesamtkonstrukt.png|center|800px]] |
| | + | |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="fps-results" name="fps-accord" type="checkbox" /> |
| | + | <label for="fps-results">Results</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | |
| | + | [[File:LMU14_GFP_pBS0K-Pspac.jpg|150px|thumb|right|Fluorescence of GFP under expression of promoter Pspac]] |
| | + | |
| | + | The mutagenesis of the Biobrick E1010 was successful and confirmed by sequencing, the thus created Biobrick is called [http://parts.igem.org/Part:BBa_K1351021 BBa_K1351021]. The fusion with the His tags were also successfully conducted and confirmed by sequencing. |
| | + | The ligation into the vector pBS0K-Pspac was confirmed via Colony PCR and the transformation into'' B. subtilis'' was conducted successfully. |
| | + | |
| | + | The intensity and spectrum of the fluorescence was measured by a TECAN Plate Reader, kindly provided by the AG Leonhardt from the LMU. |
| | + | |
| | + | The fluorescence of all of the proteins, however, proved to be not very good and often at the same level as the auto fluorescence of the ''B. subtilis'' wild type. The examination of the cells under a microscope revealed, that the promoter is not suited for this experimental set up, since the gene expression is very heterogeneous and rather low. |
| | + | |
| | + | |
| | + | |
| | + | This problem will be solved by recloning the FPs into the vector pBS1C ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K823023 BBa_K823023]) together with the xylose-inducible promoter Pxyl([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1351039 BBa_K1351039]). |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | </section> |
| | + | </div> |
| | + | </section> |
| | + | <section class="bg-color-2"> |
| | + | <div> |
| | + | </html> |
| | + | |
| | + | == Linkers == |
| | + | Linkers are short peptides used to fuse two protein domains (or even whole proteins). They are designed not to interact with the fused parts. A linker of the wrong length can lead to sterically inactivated (if the linker is too short) or instable (if it is too long) constructs. Last but not least, linkers need to exhibit a certain flexibility. We evaluated a set of linkers partially already found in the registry using FRET. |
| | + | <html> |
| | + | <section class="accordion"> |
| | + | <div> |
| | + | <input id="linker-bground" name="linker-accord" type="checkbox" /> |
| | + | <label for="linker-bground">Background</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | Förster Resonance Energy Transfer (FRET) is a phenomenon that occurs when two fluorophores with overlapping spectra are not more than 10 nanometers apart. When the donor fluorophore gets excited, and FRET happens, it does not emit all the energy as light, it transferres a part of the energy non-radiatively to the acceptor fluorophore, which starts fluorescing. The efficiency of this energy transfer decays with the sixth power of the distance between donor and acceptor. This makes FRET an ideal tool to measure or verify very short distances, like the ones between interacting proteins ''in vivo''. A common application would be tagging two supposedely interacting proteins with the FRET couple and measuring acceptor fluorescence. Obviously this technique lends itself well to the evaluation of linkers. |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="linker-design" name="linker-accord" type="checkbox" /> |
| | + | <label for="linker-design">Design</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | |
| | + | mTurquoise (donor) and mNeonGreen (acceptor) were used as FRET-couple for this study. Translational fusion constructs were created according to a modified RFC25 standard. From the registry we took the linkers [http://parts.igem.org/Part:BBa_K243004 BBa_K243004], [http://parts.igem.org/Part:BBa_K243005 BBa_K243005], [http://parts.igem.org/Part:BBa_K243006 BBa_K243006], [http://parts.igem.org/Part:BBa_K157009 BBa_K157009], [http://parts.igem.org/Part:BBa_K157013 BBa_K157013], [http://parts.igem.org/Part:BBa_K243029 BBa_K243029] and [http://parts.igem.org/Part:BBa_K243030 BBa_K243030]. We also used this method to characterize our own linkers [http://parts.igem.org/Part:BBa_K1351009 BBa_K1351009] and [http://parts.igem.org/Part:BBa_K1351035 BBa_K1351035]. For each linker a construct with mTurquise fused to it N-terminally and mNeonGreen C-terminally was created, as well as a construct where the FPs were oriented the other way around. |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="linkers-results" name="linkers-accord" type="checkbox" /> |
| | + | <label for="linkers-results">Results</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | The constructs have been created, and fluorescence has been observed. Due to time concerns, the FRET-assay has not yet been performed. |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | </section> |
| | + | </div> |
| | + | </section> |
| | + | <section> |
| | + | <div> |
| | + | </html> |
| | + | |
| | + | == ''Bacillus'' RBS collection== |
| | + | Ribosome Binding Sites (RBS) are essential for gene expression, since they are required for translation initiation. Much to our dismay, there used to be no ''Bacillus'' RBS in the registry. So we designed a broad-range RBS library for ''B. subtilis'', aiming for maximum translation initiation rate space coverage. |
| | + | |
| | + | |
| | + | <html> |
| | + | <section class="accordion"> |
| | + | <div> |
| | + | <input id="rbs-bground" name="rbs-accord" type="checkbox" /> |
| | + | <label for="rbs-bground">Background</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | RBS are short nucleotide sequences that are complemetary to the 3' end of 16S rRNA, and thus are bound by the ribosome during translation initiation. The RBS is usually found ~8 nucleotides upstream of the start codon in procaryotes. In ''B. subtilis'' the perfect consensus RBS is AAGGAGGGATA ([http://parts.igem.org/Part:BBa_K1351028 Bba_K1351028]). |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="rbs-design" name="rbs-accord" type="checkbox" /> |
| | + | <label for="rbs-design">Design</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | Using the Salis lab's [https://salis.psu.edu/software/ RBS calculator] we calculated our RBS library with the sequence ARRRRRRGATA. The 7 RBS from this collection that were submitted as BioBricks ([http://parts.igem.org/Part:BBa_K1351028 BBa_K1351028]-[http://parts.igem.org/Part:BBa_K1351034 BBa_K1351034]) are the ones that offer the best coverage of translation initiation rate space, number 1 being the optimal ''Bacillus'' RBS and number 7 being the weakest. <html> |
| | + | </article> |
| | + | </div> |
| | + | <div> |
| | + | <input id="rbs-results" name="rbs-accord" type="checkbox" /> |
| | + | <label for="rbs-results">Results</label> |
| | + | <article class="ac-small"> |
| | + | </html> |
| | + | The constructs have been created, the evaluation however is not yet finished. |
| | + | <html> |
| | + | </article> |
| | + | </div> |
| | + | </section> |
| | + | <!-- Hier kann nach dem <html> noch mal Text folgen --> |
| | + | </html> |
| | + | |
| | + | <html> |
| | + | <!-- Nach letztem Absatz! --> |
| | + | </div> |
| | + | </section> |
| | + | </section> |
| | </html> | | </html> |
| | | | |
| - | = BioBrickBox =
| |
| - | Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet. Lorem ipsum dolor sit amet, consetetur sadipscing elitr, sed diam nonumy eirmod tempor invidunt ut labore et dolore magna aliquyam erat, sed diam voluptua. At vero eos et accusam et justo duo dolores et ea rebum. Stet clita kasd gubergren, no sea takimata sanctus est Lorem ipsum dolor sit amet.
| |
| | | | |
| | {{Template:Team:LMU-Munich/Playground/footer}} | | {{Template:Team:LMU-Munich/Playground/footer}} |
| | | | |
| | <html><script>initiateNavigation("project");</script></html> | | <html><script>initiateNavigation("project");</script></html> |
 "
"