Team:Aachen/Notebook/Wetlab/September
From 2014.igem.org
Aschechtel (Talk | contribs) (→2nd) |
(→References) |
||
| (9 intermediate revisions not shown) | |||
| Line 98: | Line 98: | ||
* made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device | * made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device | ||
<center> | <center> | ||
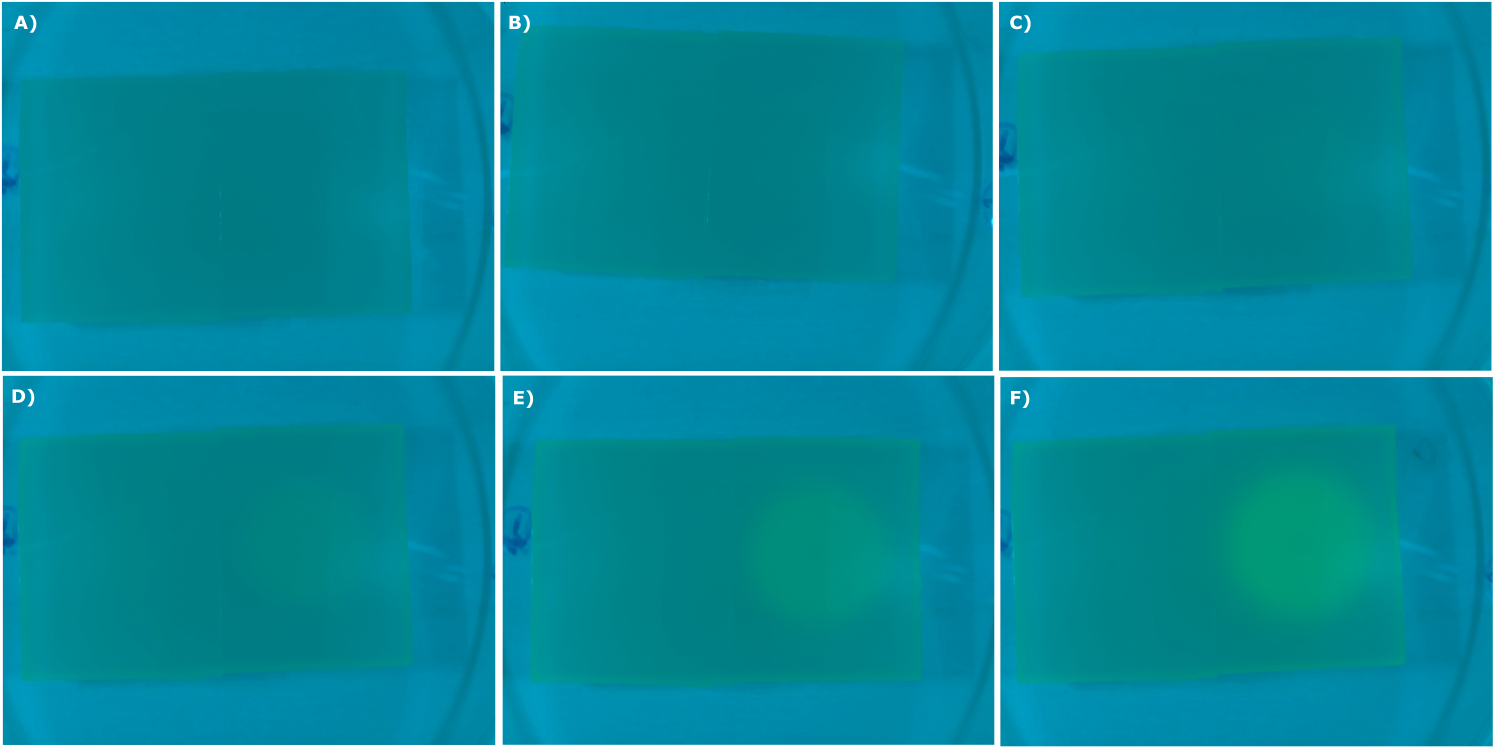
| - | {{Team:Aachen/Figure|Aachen_02_09_2014_K131026_dh5a_serie.png|title=Sensor Chips with K131026 in DH5α in LB taken with the second version of our own device|subtitle=Sensor chips with K131026 in DH5α in LB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_02_09_2014_K131026_dh5a_serie.png|title=Sensor Chips with K131026 in DH5α in LB taken with the second version of our own device|subtitle=Sensor chips with K131026 in DH5α in LB medium with 1,5% agarose, right chip induced. A) befor induction with 0.2 µl of 500 µg/ml HSL (3-oxo-C12) B) 0.5 h after induction C) 1 h after induction D) 1.5 h after induction E) 2.5 h after induction F) 3 h after induction|width=900px}} |
</center> | </center> | ||
* gel purification of vector backbones | * gel purification of vector backbones | ||
| Line 154: | Line 154: | ||
! step !! temperature [°C] !! duration | ! step !! temperature [°C] !! duration | ||
|- | |- | ||
| - | | denature || 98 || 30", | + | | denature || 98 || 30", 98°C for 10", 55°C for 30", 72°C for 2'15" |
|- | |- | ||
| denature || 98 || 10" | | denature || 98 || 10" | ||
| Line 174: | Line 174: | ||
== 5th == | == 5th == | ||
* made master plates of yesterday's transformed cells. | * made master plates of yesterday's transformed cells. | ||
| + | * As described in the article by Elling [1], Gal3 binds on the LPS of ''Pseudomonas aeruginosa''. To demonstrate this behavior several experiments were conducted. The experiments included the binding of Gal3 to ''Saccharomyces cerevisiae'', ''E.coli'', ''Pseudomonas putida'' and ''Pseudomonas aeruginosa''. The precultures were washed two times in PBS, resuspended in 10 and 50 ng/ml of YFP-Gal3 and then incubated for 1 h at room temperature. Afterwards, all samples were examined in the fluorescence microscope. As a positive control NEB10ß cells with E0030 were used. Unfortunatly, a fluorescence was not visually detectable, with exception of the positive control. | ||
== 6th == | == 6th == | ||
| Line 182: | Line 183: | ||
* made chips with K131026 in DH5α and NEB and B0015 in NEB. Images were taken every 30 min with our own device | * made chips with K131026 in DH5α and NEB and B0015 in NEB. Images were taken every 30 min with our own device | ||
<center> | <center> | ||
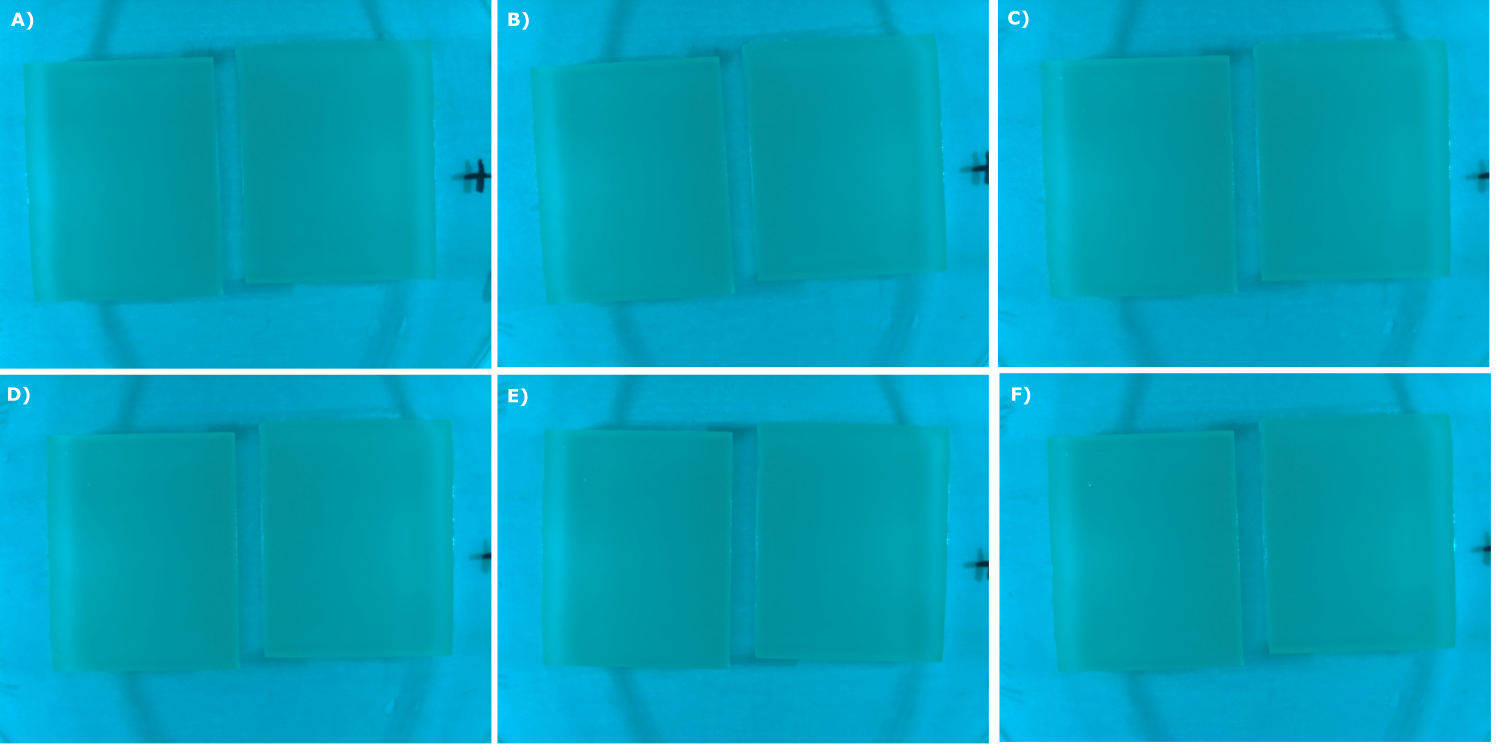
| - | {{Team:Aachen/Figure|Aachen_07_09_2014_B0015_neb_serie.png|title=Sensor Chips with B0015 in NEB (negativ control) in TB taken with the second version of our own device|subtitle=Sensor chips with B0015 in NEB in TB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_07_09_2014_B0015_neb_serie.png|title=Sensor Chips with B0015 in NEB (negativ control) in TB taken with the second version of our own device|subtitle=Sensor chips with B0015 in NEB in TB medium with 1,5% agar, right chip induced. A) befor induction with 0.2 µl of 500 µg/ml HSL (3-oxo-C12) B) 0 h after induction C) 0.5 h after induction D) 1 h after induction E) 2 h after induction F) 2.5 h after induction|width=900px}} |
</center> | </center> | ||
* purification of the following plasmids: | * purification of the following plasmids: | ||
| Line 233: | Line 234: | ||
* made chips with K131026 in DH5α and NEB and without cells. Images were taken every 30 min with our own device | * made chips with K131026 in DH5α and NEB and without cells. Images were taken every 30 min with our own device | ||
<center> | <center> | ||
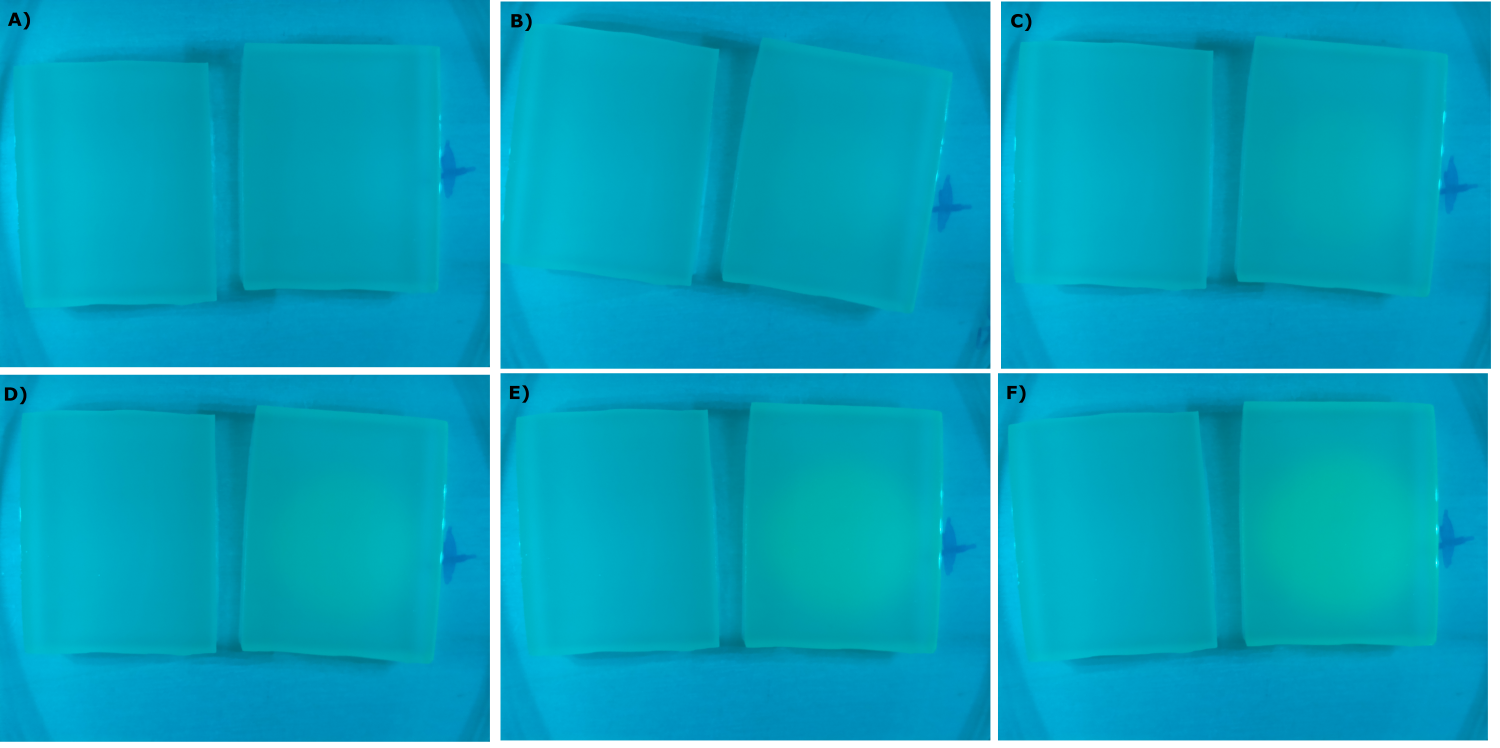
| - | {{Team:Aachen/Figure|Aachen_09_09_2014_K131026_neb_agarose_serie.png|title=Sensor Chips with K131026 in DH5α in LB|subtitle=Sensor chips with K131026 in DH5α in LB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_09_09_2014_K131026_neb_agarose_serie.png|title=Sensor Chips with K131026 in DH5α in LB|subtitle=Sensor chips with K131026 in DH5α in LB medium with 1,5% agarose, right chip induced. A) befor induction with 1 µl of 500 µg/ml HSL (3-oxo-C12) B) 1 h after induction C) 1.5 h after induction D) 2 h after induction E) 2.5 h after induction F) 3 h after induction|width=900px}} |
</center> | </center> | ||
| Line 262: | Line 263: | ||
* made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device | * made chips with K131026 in DH5α and NEB. Images were taken every 30 min with our own device | ||
<center> | <center> | ||
| - | {{Team:Aachen/Figure|Aachen_16_09_2014_K131026_neb_serie.png|title=Sensor Chips with K131026 in NEB in LB|subtitle=Sensor chips with K131026 in NEB in LB medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_16_09_2014_K131026_neb_serie.png|title=Sensor Chips with K131026 in NEB in LB|subtitle=Sensor chips with K131026 in NEB in LB medium with 1,5% agarose, right chip induced. A) 0.5 h after induction with 0.2 µl of 500 µg/ml HSL (3-oxo-C12) B) 1 h after induction C) 1.5 h after induction D) 2 h after induction E) 2.5 h after induction F) 3 h after induction|width=900px}} |
</center> | </center> | ||
| Line 272: | Line 273: | ||
* SDS page of REACh constructs with TEV and IPTG | * SDS page of REACh constructs with TEV and IPTG | ||
* over night cultures of K131026, B0015, K1319013, K1319014, K1319013 + K1319008 and K1319014 + K1319008 all in BL21 | * over night cultures of K131026, B0015, K1319013, K1319014, K1319013 + K1319008 and K1319014 + K1319008 all in BL21 | ||
| - | * tested ''Pseudomonas fluoresence'' if they are suitable for a growth experiment that is planned for our collaboration with the NEAnderLab next week. Therefore, she filled 2 500 mL flasks with 30 mL LB Pseudomonas-F medium, and inoculated each one with 1 mL culture medium of the overnight preculture. Flasks were inoculated at | + | * tested ''Pseudomonas fluoresence'' if they are suitable for a growth experiment that is planned for our collaboration with the NEAnderLab next week. Therefore, she filled 2 500 mL flasks with 30 mL LB Pseudomonas-F medium, and inoculated each one with 1 mL culture medium of the overnight preculture. Flasks were inoculated at 30°C at 250 rpm. However, after 5 hours no exponential growth could be shown (s. plot below). Thus, it was decided to use a ''E. coli'' K12 derivate strain in TB medium instead, and 30 mL of TB medium in a 500 mL flask were inoculated with ''E. coli'' DH5α cells and incubated at 37°C at 300 rpm over night. According to the [https://www.dsmz.de/catalogues/catalogue-microorganisms/groups-of-organisms-and-their-applications/strains-for-schools-and-universities.html DSMZ] ''E . coli'' K12 strain derivates, such as DH5α, are adequate for the kind of school experiment we are planning with the NEAnderLab. |
<center> | <center> | ||
| Line 279: | Line 280: | ||
== 19th == | == 19th == | ||
| - | * made flask cultures of K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 (negative control) and I20260 (positive control). iPTG was added at an OD of ~0.5. Inoculation was done via precultures in 500 ml shake flasks (50 ml filling volume). Media was always LB. Cultivation was done at | + | * made flask cultures of K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 (negative control) and I20260 (positive control). iPTG was added at an OD of ~0.5. Inoculation was done via precultures in 500 ml shake flasks (50 ml filling volume). Media was always LB. Cultivation was done at 37°C and 300 rpm. The starting OD was aimed to be 0.1. Inoculation occured directly from the precultures. Samples were taken every hour and checked for OD and fluorescence using a spectrophotometer and plate reader, respectively. |
* did plasmid preparation from the cultures of the day before (K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 and I20260). The plasmid were then be cut with EcoRI and PstI, and the results were be put on an agarose gel in order to perform a restriction test. Also plasmids of K1319013 and K1319014 will be cut with EcoRi and SpeI. K1319008 will be cut with XbaI and PstI. These will then be ligated together and then ligated into a pSB1A3 vector via the 3A assembly (vector cut with EcoRI and PstI). These constructs will be transformed into BL21 (and NEB as a backup). The created construts will be known as K1319018 (K1319013.K1319008) and K1319019 (K1319014.K1319008). | * did plasmid preparation from the cultures of the day before (K1319013, K1319013 + K1319008, K1319013 + K1319008 + iPTG, K1319014, K1319014 + K1319008, K1319014 + K1319008 + iPTG, B0015 and I20260). The plasmid were then be cut with EcoRI and PstI, and the results were be put on an agarose gel in order to perform a restriction test. Also plasmids of K1319013 and K1319014 will be cut with EcoRi and SpeI. K1319008 will be cut with XbaI and PstI. These will then be ligated together and then ligated into a pSB1A3 vector via the 3A assembly (vector cut with EcoRI and PstI). These constructs will be transformed into BL21 (and NEB as a backup). The created construts will be known as K1319018 (K1319013.K1319008) and K1319019 (K1319014.K1319008). | ||
| Line 285: | Line 286: | ||
* made precultures of the master plates from the day before (K1319008, K1319013, K1319015 and pSBX1A3 with Gal3). | * made precultures of the master plates from the day before (K1319008, K1319013, K1319015 and pSBX1A3 with Gal3). | ||
| - | * also inoculated 4 cultures for the further testing of the OD/F | + | * also inoculated 4 cultures for the further testing of the OD/F Device (the F part). The cultures are 2 shake flasks of I20260 and 2 shake flasks of B0015. |
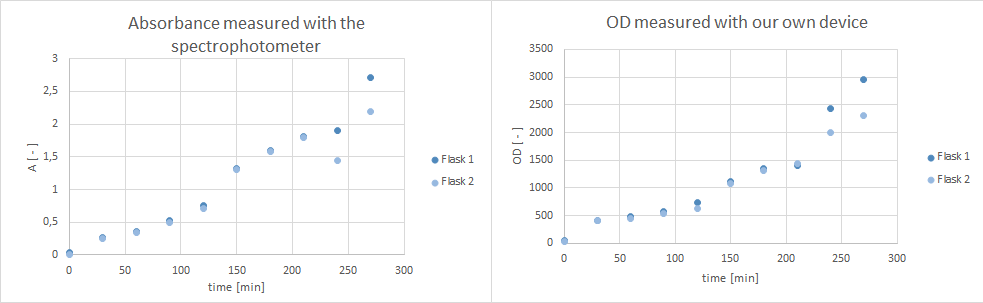
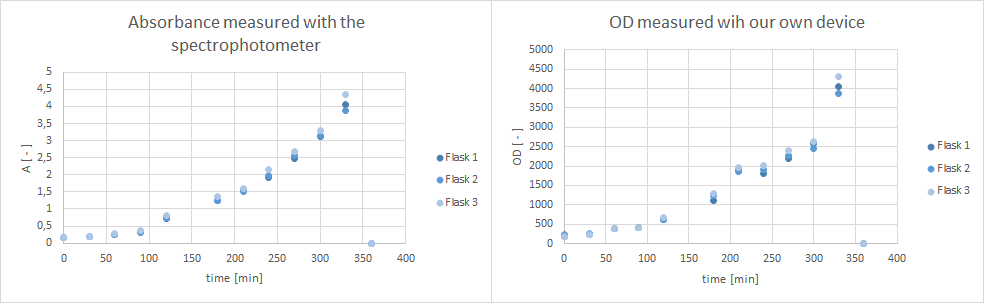
| - | * Furthermore, did a growth experiment with | + | * Furthermore, did a growth experiment with DH5α for the NEAnderLab school experiment. 3 500 mL shake flasks were filled with 50 mL TB medium, and inoculated to an OD of 1.5 with the overnight preculture. Samples were taken every 30 minutes and tested for OD using our own device as well as the spectrophotometer. The resulting growth curve is shown below. we concluded that the growth was fast enough for these growth conditions to be used for the school experiment on the 24th. |
<center> | <center> | ||
| Line 295: | Line 296: | ||
* made chips with K1319013 + K1319008, K1319014 + K1319008, K1319013, K1319014, B0015 and K131026. Images were taken every 30 minutes with our own device. | * made chips with K1319013 + K1319008, K1319014 + K1319008, K1319013, K1319014, B0015 and K131026. Images were taken every 30 minutes with our own device. | ||
| - | * tested our OD/F | + | * tested our OD/F Device with a dilution test. Samples were checked with the spectrophotometer (OD), our OD/F Device (fluorescence) and platereader (fluorescence). |
* made two SDS gels. | * made two SDS gels. | ||
| Line 344: | Line 345: | ||
| G00100_Alternative || G00101_Alternative || water || --- || ??? | | G00100_Alternative || G00101_Alternative || water || --- || ??? | ||
|} | |} | ||
| - | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_1.png|title=Gradient PCR 1|subtitle=the primers were G00100_Alternative and G00101_Alternative and they worked well at all temperatures from 55- | + | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_1.png|title=Gradient PCR 1|subtitle=the primers were G00100_Alternative and G00101_Alternative and they worked well at all temperatures from 55-65°C.|width=800px}} |
</center> | </center> | ||
| Line 360: | Line 361: | ||
| G00100_Alternative || I746916_check_R || water || --- || ??? | | G00100_Alternative || I746916_check_R || water || --- || ??? | ||
|} | |} | ||
| - | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_2.png|title=Gradient PCR 2|subtitle=the primers were G00100_Alternative and I746916_check_R and they worked well at all temperatures from 55- | + | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_2.png|title=Gradient PCR 2|subtitle=the primers were G00100_Alternative and I746916_check_R and they worked well at all temperatures from 55-65°C. Apparently the K1319008 template contained I746916.|width=800px}} |
</center> | </center> | ||
| Line 376: | Line 377: | ||
| G00100_Alternative || K1319004_check_R || water || --- || ??? | | G00100_Alternative || K1319004_check_R || water || --- || ??? | ||
|} | |} | ||
| - | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_3.png|title=Gradient PCR 3|subtitle=The primers were G00100_Alternative and K1319004_check_R and they worked well at all temperatures from 60- | + | {{Team:Aachen/Figure|Aachen_14-09-26_gradientPCR_3.png|title=Gradient PCR 3|subtitle=The primers were G00100_Alternative and K1319004_check_R and they worked well at all temperatures from 60-68°C. To our disappointment, the K1319008 template did not contain K1319004. It is unclear why the 5 bands of K1319008 and I746916 look different.|width=800px}} |
</center> | </center> | ||
The results of these three PCRs are: | The results of these three PCRs are: | ||
# KAPA2G Fast ReadyMix worked well | # KAPA2G Fast ReadyMix worked well | ||
| - | # all three primers work well at > | + | # all three primers work well at >65°C annealing temperature |
# K1319008 template contained I746916 instead of the intended K1319004 ORF | # K1319008 template contained I746916 instead of the intended K1319004 ORF | ||
| - | It was concluded that a similar check PCR with | + | It was concluded that a similar check PCR with 65°C annealing temperature will be done on all plasmids and cryos of K1319008. |
== 27th == | == 27th == | ||
| Line 423: | Line 424: | ||
* made cryo cultures and plasmid preparation of K1319010, K1319011, K1319012, K1319021 and K1319042. We determined the contentration of plasmids and made did a restriction digest of K1319010, K1319011, K1319012, pSB1C3, K1319021, K1319013 and K1319014, followed by a ligation in K1319010.pSB1C3, K1319011.pSB1C3, K1319012.pSB1C3, K1319021.K1319013.pSB1A3 and K1319021.K1319013.pSB1A3. All constructs were transformed into ''E. coli'' NEB 10ß. | * made cryo cultures and plasmid preparation of K1319010, K1319011, K1319012, K1319021 and K1319042. We determined the contentration of plasmids and made did a restriction digest of K1319010, K1319011, K1319012, pSB1C3, K1319021, K1319013 and K1319014, followed by a ligation in K1319010.pSB1C3, K1319011.pSB1C3, K1319012.pSB1C3, K1319021.K1319013.pSB1A3 and K1319021.K1319013.pSB1A3. All constructs were transformed into ''E. coli'' NEB 10ß. | ||
| - | * prepared 3 500 mL flasks with 30 mL LB medium which were inoculated with a ''Pseudomonas putida'' strain. The cells were cultured over night at | + | * prepared 3 500 mL flasks with 30 mL LB medium which were inoculated with a ''Pseudomonas putida'' strain. The cells were cultured over night at 28°C and ~300 rpm. The cultures are supposed to be used to test our OD device. |
== 30th == | == 30th == | ||
| Line 515: | Line 516: | ||
</center> | </center> | ||
</html> | </html> | ||
| + | |||
| + | == References == | ||
| + | Kupper C.E, Böcker S, Liu H, et al. Fluorescent SNAP-tag galectin fusion proteins as novel tools in glycobiology. Curr Pharm Des. 2013;19(30):5457-67. Available at: http://www.ncbi.nlm.nih.gov/pubmed/23431989. | ||
| + | |||
| + | |||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Latest revision as of 23:35, 17 October 2014
|
 "
"