Team:ETH Zurich/lab/sequences
From 2014.igem.org
| (74 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:ETH_Zurich/tpl/head|Sequences}} | {{:Team:ETH_Zurich/tpl/head|Sequences}} | ||
<html><article></html> | <html><article></html> | ||
| + | |||
| + | The plasmid sequences can be accessed by clicking on the plasmid name (e.g. [https://static.igem.org/mediawiki/2014/f/f6/ETH2014_piG0040.txt piG0040]) or the plasmid picture. | ||
| + | |||
| + | Construct types: | ||
| + | |||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#Regulator_Constructs|Regulator Constructs]] | ||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#Producer_Constructs|Producer Constructs]] | ||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#Sensor Constructs|Sensor Constructs]] | ||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#BUFFER_Gate_Construct|BUFFER Gate Construct]] | ||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#Integrase_Reporter_Construct|Integrase Reporter Construct]] | ||
| + | |||
| + | [[Team:ETH_Zurich/lab/sequences#Combined_Sensor_and_Producer_Constructs|Combined Sensor and Producer Constructs]] | ||
| + | |||
| + | |||
==Regulator Constructs== | ==Regulator Constructs== | ||
| Line 7: | Line 26: | ||
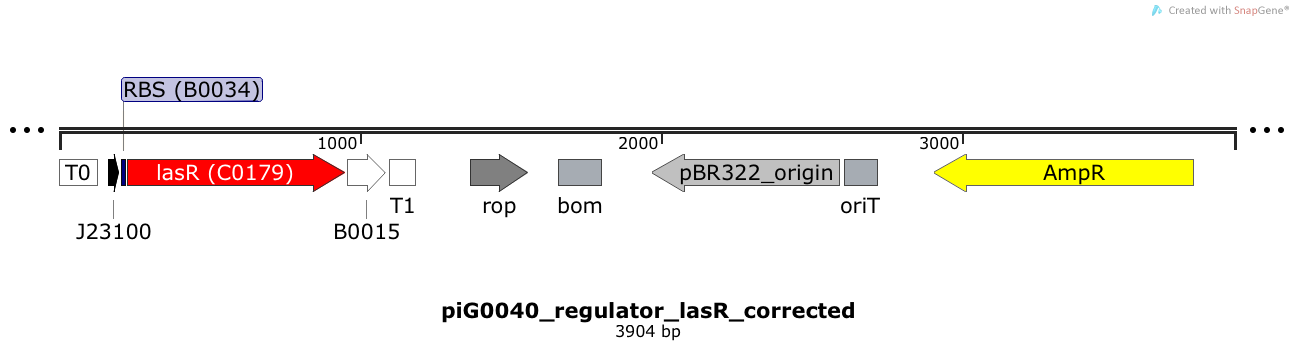
[https://static.igem.org/mediawiki/2014/f/f6/ETH2014_piG0040.txt piG0040] | [https://static.igem.org/mediawiki/2014/f/f6/ETH2014_piG0040.txt piG0040] | ||
| - | LasR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] | + | LasR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LasR bound to 3OC12-HSL induces the expression of genes under the control of pLasR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | [[File:ETH2014_piG0040_Map.png|link=https://static.igem.org/mediawiki/2014/f/f6/ETH2014_piG0040.txt]] | ||
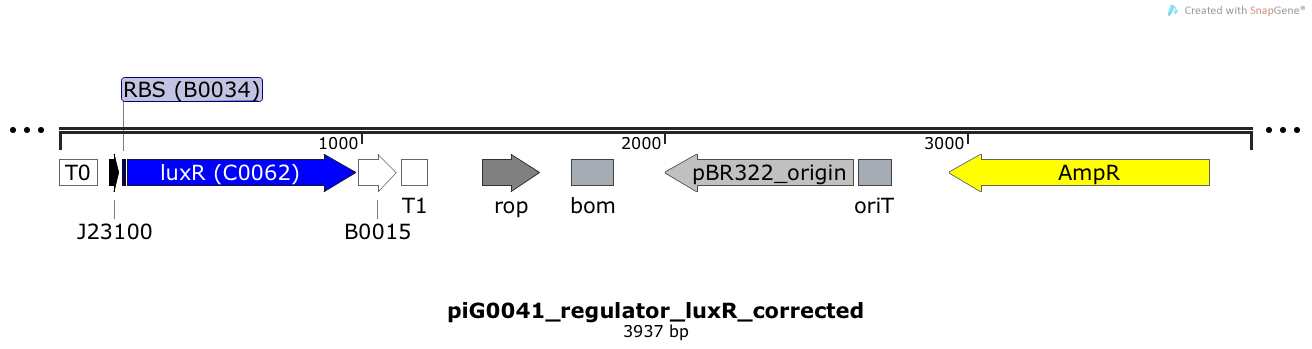
[https://static.igem.org/mediawiki/2014/2/2c/ETH2014_piG0041.txt piG0041] | [https://static.igem.org/mediawiki/2014/2/2c/ETH2014_piG0041.txt piG0041] | ||
| - | LuxR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] | + | LuxR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0041_Map.png|link=https://static.igem.org/mediawiki/2014/2/2c/ETH2014_piG0041.txt]] | ||
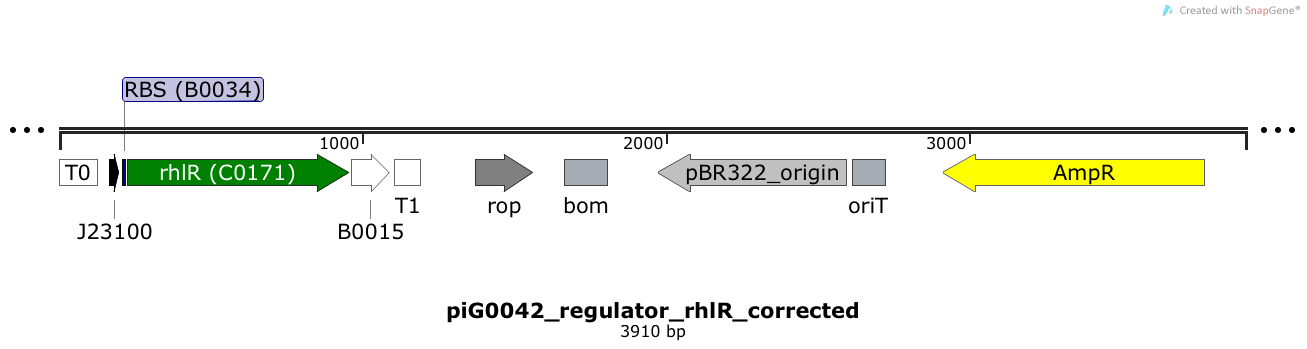
[https://static.igem.org/mediawiki/2014/d/db/ETH2014_piG0042.txt piG0042] | [https://static.igem.org/mediawiki/2014/d/db/ETH2014_piG0042.txt piG0042] | ||
| - | RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] | + | RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. RhlR bound to C4-HSL induces the expression of genes under the control of pRhlR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0042_Map.png|link=https://static.igem.org/mediawiki/2014/d/db/ETH2014_piG0042.txt]] | ||
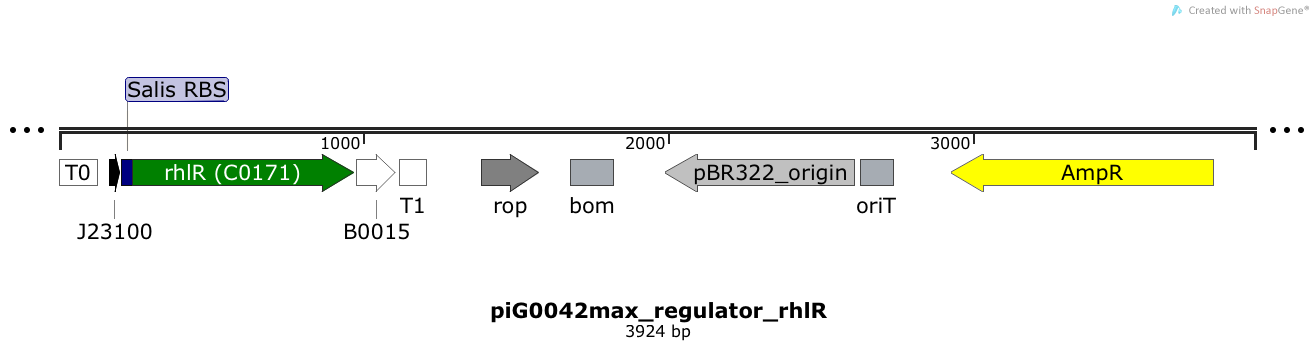
[https://static.igem.org/mediawiki/2014/c/ce/ETH2014_piG0042max.txt piG0042max] | [https://static.igem.org/mediawiki/2014/c/ce/ETH2014_piG0042max.txt piG0042max] | ||
| - | RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] | + | RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by an RBS optimised for RhlR ([https://salis.psu.edu/software/forward RBS calculator]). RhlR bound to C4-HSL induces the expression of genes under the control of pRhlR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0042max_Map.png|link=https://static.igem.org/mediawiki/2014/c/ce/ETH2014_piG0042max.txt]] | ||
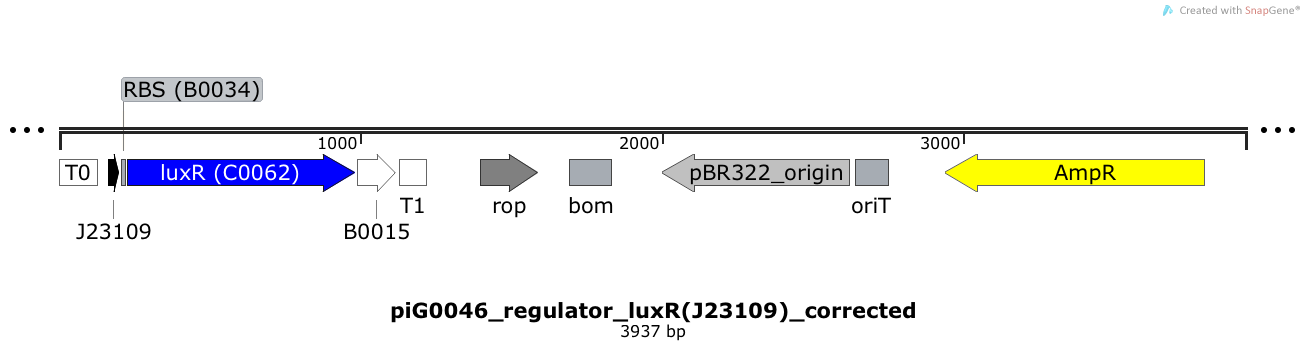
[https://static.igem.org/mediawiki/2014/1/18/ETH2014_piG0046.txt piG0046] | [https://static.igem.org/mediawiki/2014/1/18/ETH2014_piG0046.txt piG0046] | ||
| - | LuxR is expressed under the weak constitutive promoter [http://parts.igem.org/Part:BBa_J23109 BBa_J23109] | + | LuxR is expressed under the weak constitutive promoter [http://parts.igem.org/Part:BBa_J23109 BBa_J23109] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0046_Map.png|link=https://static.igem.org/mediawiki/2014/1/18/ETH2014_piG0046.txt]] | ||
| + | |||
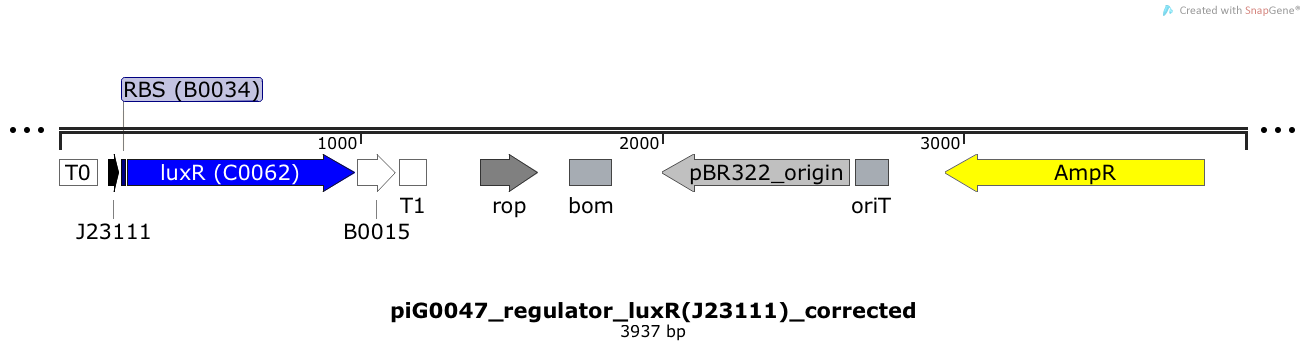
[https://static.igem.org/mediawiki/2014/1/1b/ETH2014_piG0047.txt piG0047] | [https://static.igem.org/mediawiki/2014/1/1b/ETH2014_piG0047.txt piG0047] | ||
| - | LuxR is expressed under the medium strong constitutive promoter [http://parts.igem.org/Part:BBa_J23111 BBa_J23111 ] | + | LuxR is expressed under the medium strong constitutive promoter [http://parts.igem.org/Part:BBa_J23111 BBa_J23111 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20<sup>[[Team:ETH_Zurich/project/references#refBolivar|[15]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0047_Map.png|link=https://static.igem.org/mediawiki/2014/1/1b/ETH2014_piG0047.txt]] | ||
| Line 37: | Line 74: | ||
==Producer Constructs== | ==Producer Constructs== | ||
| + | |||
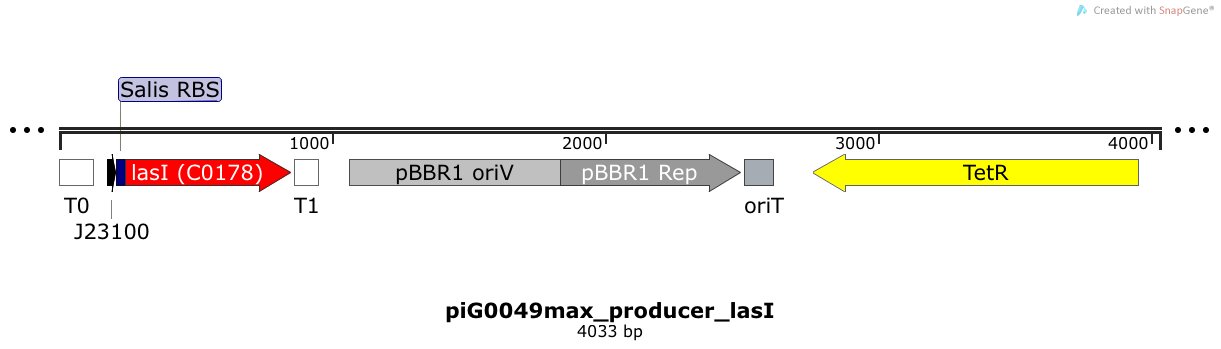
[https://static.igem.org/mediawiki/2014/4/43/ETH2014_piG0049.txt piG0049] | [https://static.igem.org/mediawiki/2014/4/43/ETH2014_piG0049.txt piG0049] | ||
| - | LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LasI produces the quorum sensing molecule 3OC12-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized <sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0049_Map.png|link=https://static.igem.org/mediawiki/2014/4/43/ETH2014_piG0049.txt]] | ||
[https://static.igem.org/mediawiki/2014/b/bc/ETH2014_piG0049max.txt piG0049max] | [https://static.igem.org/mediawiki/2014/b/bc/ETH2014_piG0049max.txt piG0049max] | ||
| - | LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for LasI ([https://salis.psu.edu/software/forward RBS calculator]). LasI produces the quorum sensing molecule 3OC12-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized <sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0049max_Map.png|link=https://static.igem.org/mediawiki/2014/b/bc/ETH2014_piG0049max.txt]] | ||
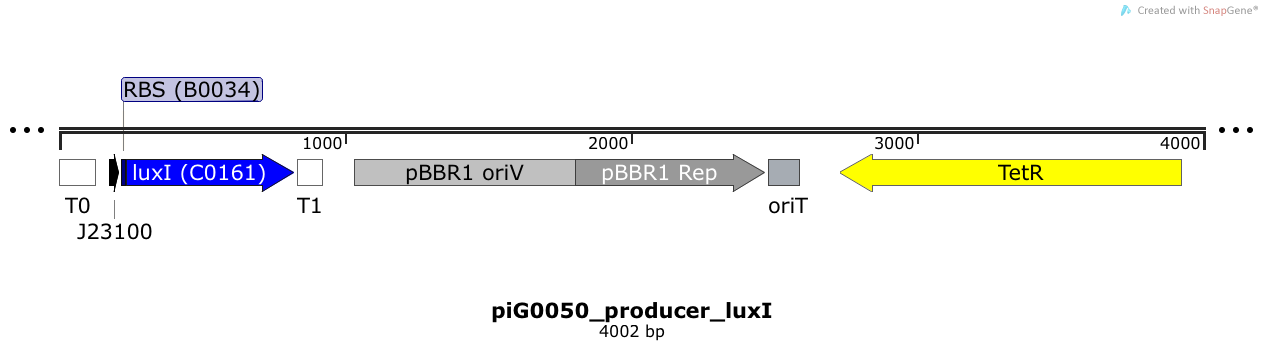
[https://static.igem.org/mediawiki/2014/4/49/ETH2014_piG0050.txt piG0050] | [https://static.igem.org/mediawiki/2014/4/49/ETH2014_piG0050.txt piG0050] | ||
| - | LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxI produces the quorum sensing molecule 3OC6-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized<sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | [[File:ETH2014_piG0050_Map.png|link=https://static.igem.org/mediawiki/2014/4/49/ETH2014_piG0050.txt]] | ||
[https://static.igem.org/mediawiki/2014/5/50/ETH2014_piG0050max.txt piG0050max] | [https://static.igem.org/mediawiki/2014/5/50/ETH2014_piG0050max.txt piG0050max] | ||
| - | LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for LuxI ([https://salis.psu.edu/software/forward RBS calculator]). LuxI produces the quorum sensing molecule 3OC6-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized<sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | |||
| + | [[File:ETH2014_piG0050max_Map.png|link=https://static.igem.org/mediawiki/2014/5/50/ETH2014_piG0050max.txt]] | ||
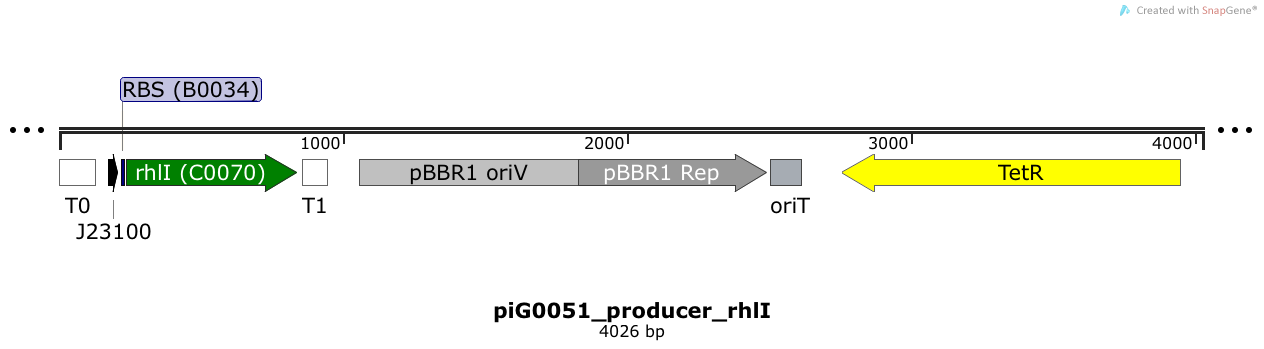
[https://static.igem.org/mediawiki/2014/0/00/ETH2014_piG0051.txt piG0051] | [https://static.igem.org/mediawiki/2014/0/00/ETH2014_piG0051.txt piG0051] | ||
| - | RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. RhlI produces the quorum sensing molecule C4-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized<sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | [[File:ETH2014_piG0051_Map.png|link=https://static.igem.org/mediawiki/2014/0/00/ETH2014_piG0051.txt]] | ||
| + | |||
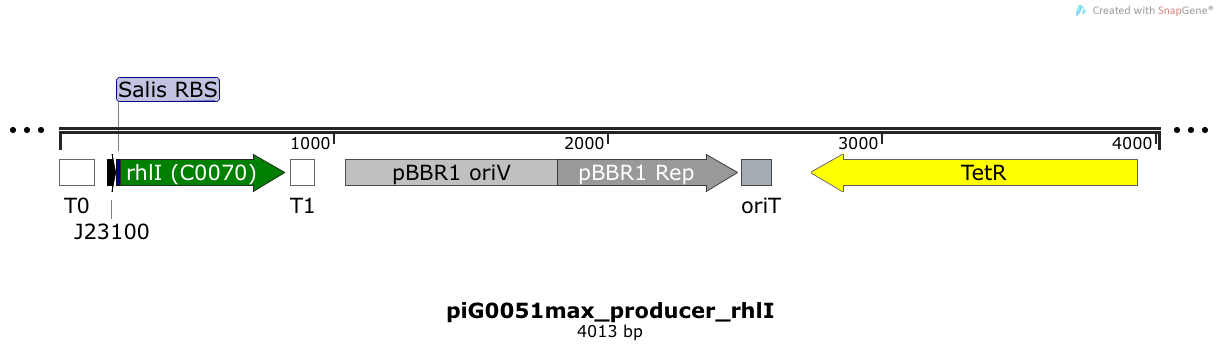
[https://static.igem.org/mediawiki/2014/9/94/ETH2014_piG0051max.txt piG0051max] | [https://static.igem.org/mediawiki/2014/9/94/ETH2014_piG0051max.txt piG0051max] | ||
| - | RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] | + | RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for RhlI ([https://salis.psu.edu/software/forward RBS calculator]). RhlI produces the quorum sensing molecule C4-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized<sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup>. |
| + | |||
| + | [[File:ETH2014_piG0051max_Map.png|link=https://static.igem.org/mediawiki/2014/9/94/ETH2014_piG0051max.txt]] | ||
| + | |||
| + | ==Sensor Constructs== | ||
| + | |||
| + | |||
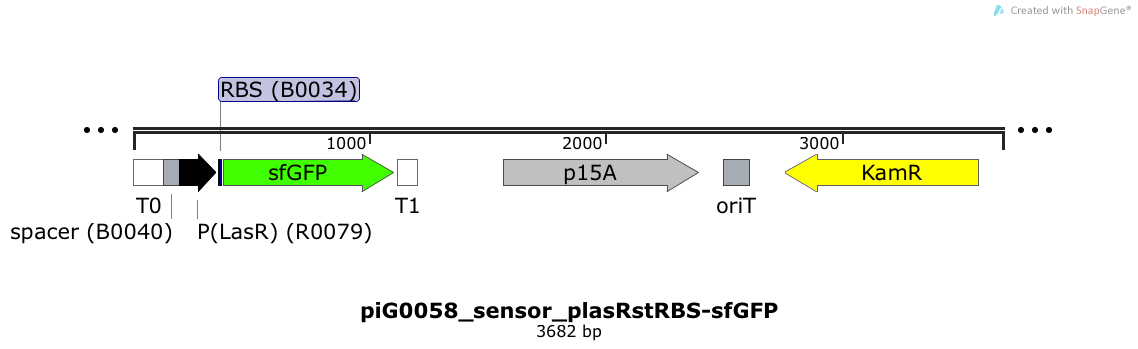
| + | [https://static.igem.org/mediawiki/2014/a/a7/ETH2014_piG0058_sensor_plasRstRBS-sfGFP.txt piG0058] | ||
| + | |||
| + | Expression of sfGFP is induced when LasR bound to 3OC12-HSL bind to pLasR. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0058 sensor plasRstRBS-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/a/a7/ETH2014_piG0058_sensor_plasRstRBS-sfGFP.txt]] | ||
| + | |||
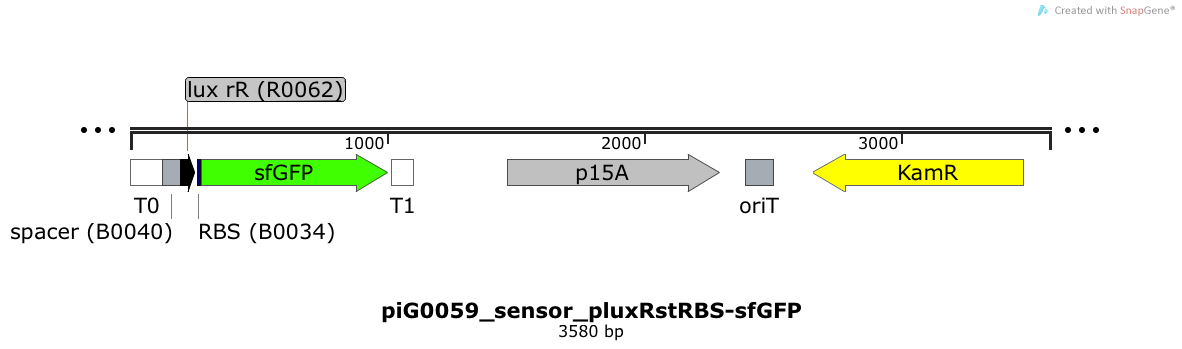
| + | [https://static.igem.org/mediawiki/2014/0/0f/ETH2014_piG0059_sensor_pluxRstRBS-sfGFP.txt piG0059] | ||
| + | |||
| + | Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0059 sensor pluxRstRBS-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/0/0f/ETH2014_piG0059_sensor_pluxRstRBS-sfGFP.txt]] | ||
| + | |||
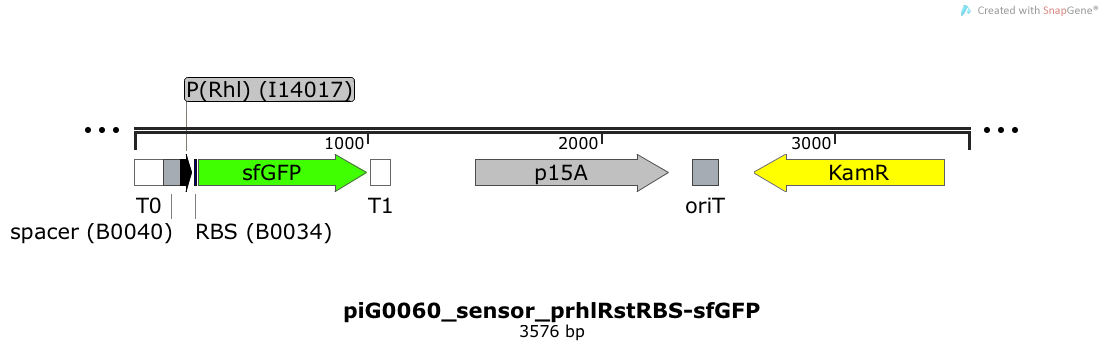
| + | [https://static.igem.org/mediawiki/2014/2/22/PiG0060_sensor_prhlRstRBS-sfGFP.txt piG0060] | ||
| + | |||
| + | Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0060 sensor prhlRstRBS-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/2/22/PiG0060_sensor_prhlRstRBS-sfGFP.txt]] | ||
| + | |||
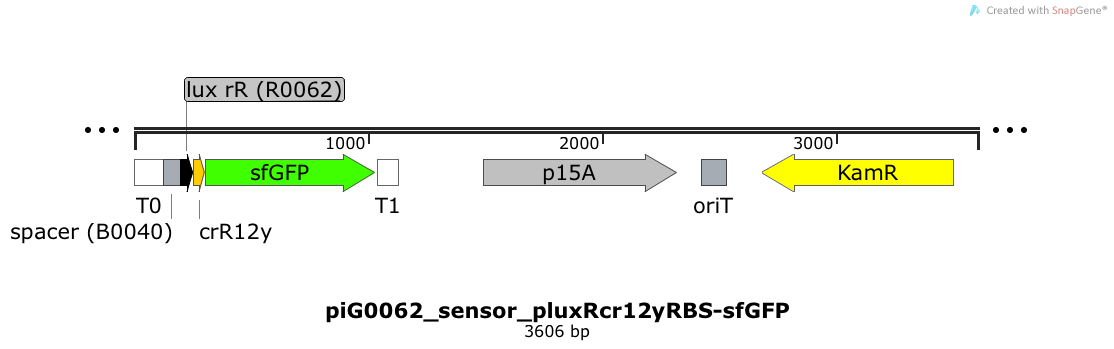
| + | [https://static.igem.org/mediawiki/2014/2/2c/PiG0062_sensor_pluxRcr12yRBS-sfGFP.txt piG0062] | ||
| + | |||
| + | Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0062 sensor pluxRcr12yRBS-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/2/2c/PiG0062_sensor_pluxRcr12yRBS-sfGFP.txt]] | ||
| + | |||
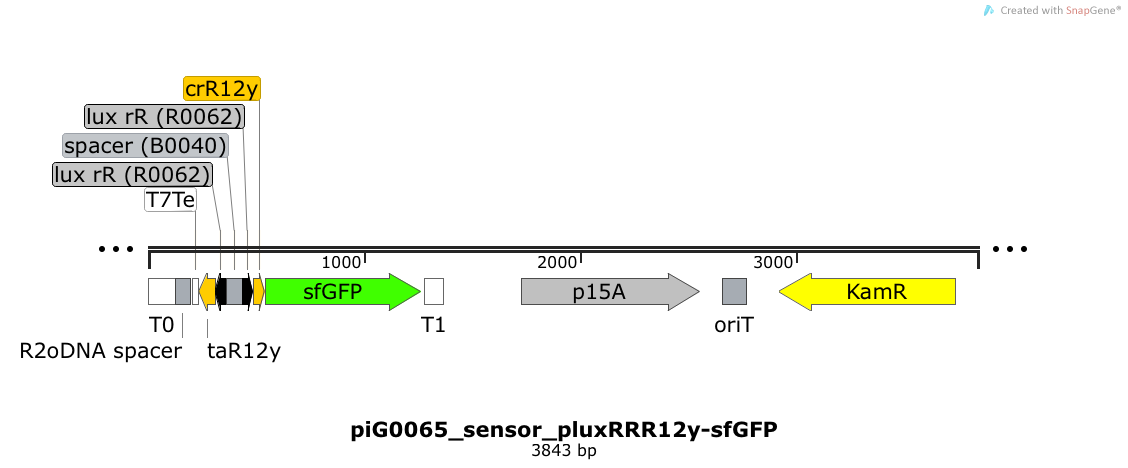
| + | [https://static.igem.org/mediawiki/2014/d/da/ETH2014_piG0065_sensor_pluxRRR12y-sfGFP.txt piG0065] | ||
| + | |||
| + | Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0065 sensor pluxRRR12y-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/d/da/ETH2014_piG0065_sensor_pluxRRR12y-sfGFP.txt]] | ||
| + | |||
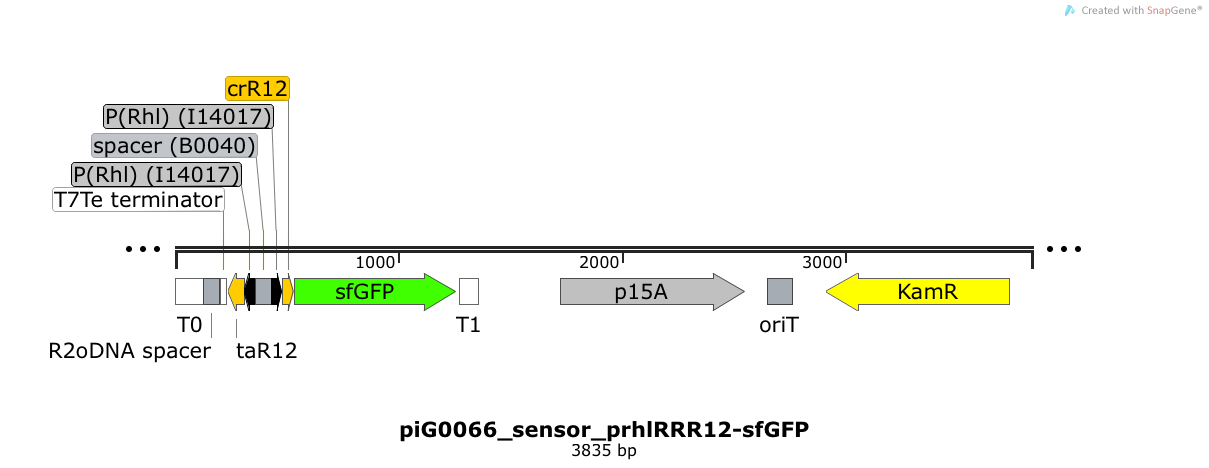
| + | [https://static.igem.org/mediawiki/2014/2/24/ETH2014_piG0066_sensor_prhlRRR12-sfGFP.txt piG0066] | ||
| + | |||
| + | Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The cis-repressive element (crR12) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0066 sensor prhlRRR12-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/2/24/ETH2014_piG0066_sensor_prhlRRR12-sfGFP.txt]] | ||
| + | |||
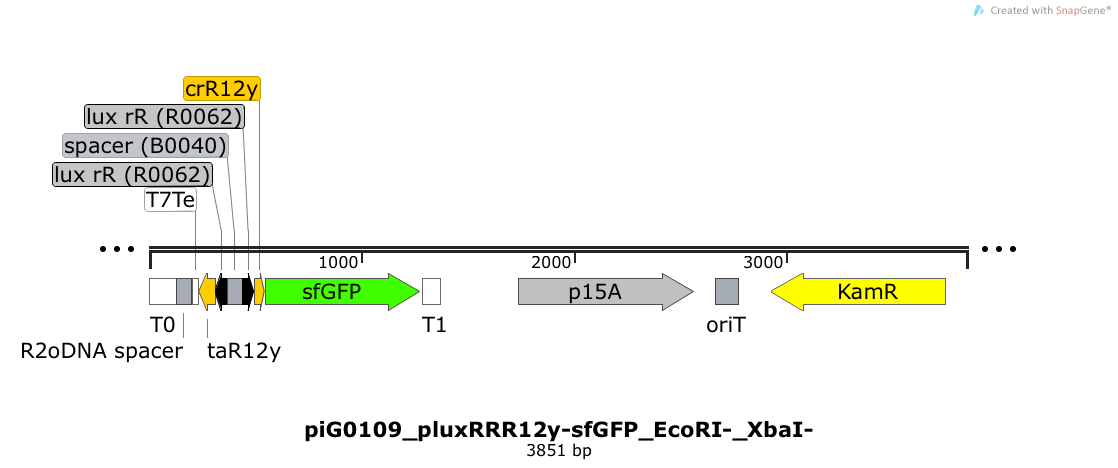
| + | [https://static.igem.org/mediawiki/2014/9/95/ETH2014_piG0109_pluxRRR12y-sfGFP_EcoRI-_XbaI-.txt piG0109] | ||
| + | |||
| + | Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. PiG0109 is a derivate of piG0065 where the restriction sites EcoRI and XbaI have been removed. Thus, the two constructs slightly differ in the sequence of the 3'-end of the trans-activating element and in the sequence of the 5'-end of the cis-repressive element. | ||
| + | |||
| + | [[File:ETH2014 piG0109 pluxRRR12y-sfGFP EcoRI- XbaI- Map.png|link=https://static.igem.org/mediawiki/2014/9/95/ETH2014_piG0109_pluxRRR12y-sfGFP_EcoRI-_XbaI-.txt]] | ||
| + | |||
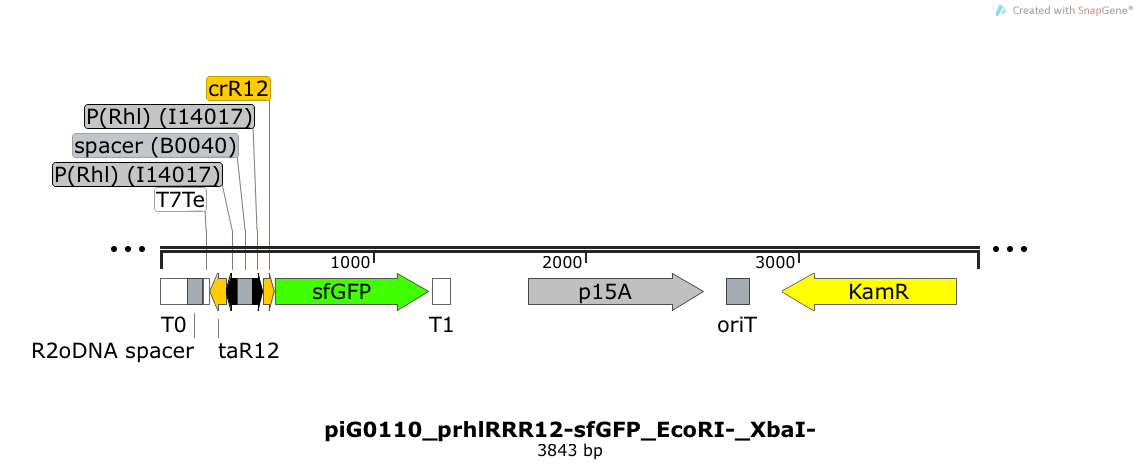
| + | [https://static.igem.org/mediawiki/2014/f/f9/ETH2014_piG0110_prhlRRR12-sfGFP_EcoRI-_XbaI-.txt piG0110] | ||
| + | |||
| + | Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The cis-repressive element (crR12) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. PiG0110 is a derivate of piG0066 where the restriction sites EcoRI and XbaI have been removed. Thus, the two constructs slightly differ in the sequence of the 3'-end of the trans-activating element and in the sequence of the 5'-end of the cis-repressive element. | ||
| + | |||
| + | [[File:ETH2014 piG0110 prhlRRR12-sfGFP EcoRI- XbaI- Map.png|link=https://static.igem.org/mediawiki/2014/f/f9/ETH2014_piG0110_prhlRRR12-sfGFP_EcoRI-_XbaI-.txt]] | ||
| + | |||
| + | ==BUFFER Gate Construct== | ||
| + | |||
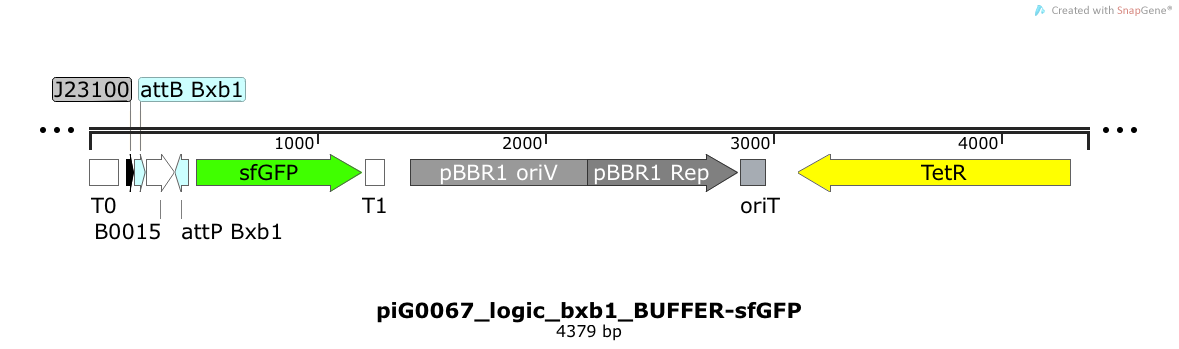
| + | [https://static.igem.org/mediawiki/2014/5/59/ETH2014_piG0067_logic_bxb1_BUFFER-sfGFP.txt piG0067] | ||
| + | |||
| + | The directional terminator [http://parts.igem.org/Part:BBa_B0015 BBa_B0015] blocks transcription of sfGFP under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100]. Only if the integrase Bxb1 flips B0015 between the attP and the attB sites, transcription of sfGFP is possible. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized<sup>[[Team:ETH_Zurich/project/references#refLennen|[16]]]</sup> | ||
| + | |||
| + | |||
| + | [[File:ETH2014 piG0067 logic bxb1 BUFFER-sfGFP Map.png|link=https://static.igem.org/mediawiki/2014/5/59/ETH2014_piG0067_logic_bxb1_BUFFER-sfGFP.txt]] | ||
| + | |||
| + | ==Integrase Reporter Construct== | ||
| + | |||
| + | |||
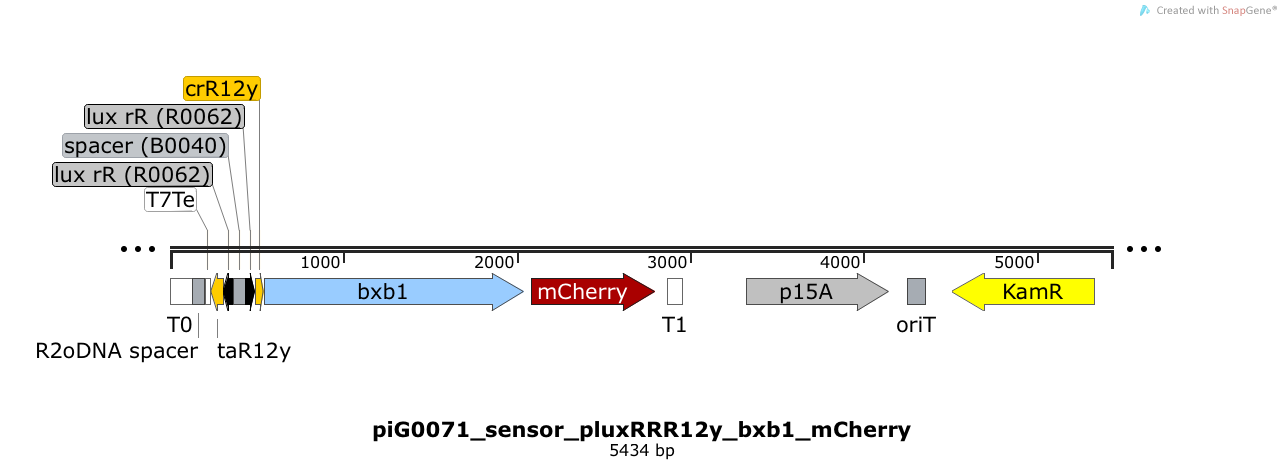
| + | [https://static.igem.org/mediawiki/2014/1/1d/ETH2014_piG0071_sensor_pluxRRR12y_bxb1_mCherry.txt piG0071] | ||
| + | |||
| + | Expression of the integrase Bxb1 and the fluorophore mCherry is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of Bxb1 and mCherry, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0071 sensor pluxRRR12y bxb1 mCherry Map.png|link=https://static.igem.org/mediawiki/2014/1/1d/ETH2014_piG0071_sensor_pluxRRR12y_bxb1_mCherry.txt]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ==Combined Sensor and Producer Constructs== | ||
| + | |||
| + | |||
| + | [https://static.igem.org/mediawiki/2014/f/f5/ETH2014_piG0096_pluxRRR12y-sfGFP-maxRBS-luxI.txt piG0096] | ||
| + | |||
| + | Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of the succeeding gene, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. The expression level of LuxI is increased by an RBS optimised for LuxI ([https://salis.psu.edu/software/forward RBS calculator]). A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0096 pluxRRR12y-sfGFP-maxRBS-luxI Map.png|link=https://static.igem.org/mediawiki/2014/f/f5/ETH2014_piG0096_pluxRRR12y-sfGFP-maxRBS-luxI.txt]] | ||
| + | |||
| + | [https://static.igem.org/mediawiki/2014/f/fc/ETH2014_piG0097_pluxRRR12y-sfGFP-stRBS-luxI.txt piG0097] | ||
| + | |||
| + | Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of the succeeding gene, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a [https://2014.igem.org/Team:ETH_Zurich/expresults#Riboregulators riboregulator] that decreases leakiness of pLuxR. The expression level of LuxI is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. A biologically neutral spacer sequence was designed using the web application R2oDNA<sup>[[Team:ETH_Zurich/project/references#refR2oDNA|[34]]]</sup>. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014 piG0097 pluxRRR12y-sfGFP-stRBS-luxI Map.png|link=https://static.igem.org/mediawiki/2014/f/fc/ETH2014_piG0097_pluxRRR12y-sfGFP-stRBS-luxI.txt]] | ||
| + | |||
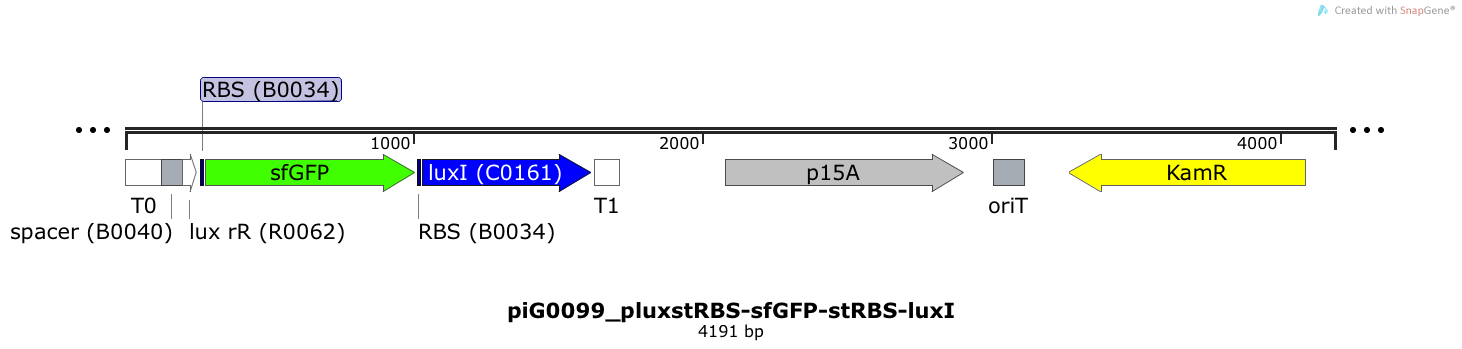
| + | [https://static.igem.org/mediawiki/2014/6/69/ETH2014_piG0099_pluxstRBS-sfGFP-stRBS-luxI.txt piG0099] | ||
| + | |||
| + | Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The expression level of sfGFP and LuxI is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. The p15A is present at a copy number of approximately 15 to 25<sup>[[Team:ETH_Zurich/project/references#refChang|[35]]]</sup>. | ||
| + | |||
| + | [[File:ETH2014_piG0099_pluxstRBS-sfGFP-stRBS-luxI_Map.png|link=https://static.igem.org/mediawiki/2014/6/69/ETH2014_piG0099_pluxstRBS-sfGFP-stRBS-luxI.txt]] | ||
| + | |||
<html></article></html> | <html></article></html> | ||
{{:Team:ETH_Zurich/tpl/foot}} | {{:Team:ETH_Zurich/tpl/foot}} | ||
Latest revision as of 03:28, 18 October 2014
Sequences
The plasmid sequences can be accessed by clicking on the plasmid name (e.g. piG0040) or the plasmid picture.
Construct types:
Combined Sensor and Producer Constructs
Regulator Constructs
LasR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LasR bound to 3OC12-HSL induces the expression of genes under the control of pLasR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
LuxR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. RhlR bound to C4-HSL induces the expression of genes under the control of pRhlR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
RhlR is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100] while the expression level is influenced by an RBS optimised for RhlR (RBS calculator). RhlR bound to C4-HSL induces the expression of genes under the control of pRhlR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
LuxR is expressed under the weak constitutive promoter [http://parts.igem.org/Part:BBa_J23109 BBa_J23109] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
LuxR is expressed under the medium strong constitutive promoter [http://parts.igem.org/Part:BBa_J23111 BBa_J23111 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxR bound to 3OC6-HSL induces the expression of genes under the control of pLuxR. The plasmid pBR322 and its derivatives have a copy number of 15 to 20[15].
Producer Constructs
LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LasI produces the quorum sensing molecule 3OC12-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized [16].
LasI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for LasI (RBS calculator). LasI produces the quorum sensing molecule 3OC12-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized [16].
LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. LuxI produces the quorum sensing molecule 3OC6-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized[16].
LuxI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for LuxI (RBS calculator). LuxI produces the quorum sensing molecule 3OC6-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized[16].
RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. RhlI produces the quorum sensing molecule C4-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized[16].
RhlI is expressed under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100 ] while the expression level is increased by an RBS optimised for RhlI (RBS calculator). RhlI produces the quorum sensing molecule C4-HSL. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized[16].
Sensor Constructs
Expression of sfGFP is induced when LasR bound to 3OC12-HSL bind to pLasR. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The cis-repressive element (crR12) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35]. PiG0109 is a derivate of piG0065 where the restriction sites EcoRI and XbaI have been removed. Thus, the two constructs slightly differ in the sequence of the 3'-end of the trans-activating element and in the sequence of the 5'-end of the cis-repressive element.
Expression of sfGFP is induced when RhlR bound to 4C-HSL bind to pRhlR. The cis-repressive element (crR12) inhibits the translation of sfGFP, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35]. PiG0110 is a derivate of piG0066 where the restriction sites EcoRI and XbaI have been removed. Thus, the two constructs slightly differ in the sequence of the 3'-end of the trans-activating element and in the sequence of the 5'-end of the cis-repressive element.
BUFFER Gate Construct
The directional terminator [http://parts.igem.org/Part:BBa_B0015 BBa_B0015] blocks transcription of sfGFP under the strong constitutive promoter [http://parts.igem.org/Part:BBa_J23100 BBa_J23100]. Only if the integrase Bxb1 flips B0015 between the attP and the attB sites, transcription of sfGFP is possible. The pBBR1 origin is present at a copy number of approximately 5, however, the origin is poorly characterized[16]
Integrase Reporter Construct
Expression of the integrase Bxb1 and the fluorophore mCherry is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of Bxb1 and mCherry, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35].
Combined Sensor and Producer Constructs
Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of the succeeding gene, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. The expression level of LuxI is increased by an RBS optimised for LuxI (RBS calculator). A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The cis-repressive element (crR12y) inhibits the translation of the succeeding gene, since the RBS is blocked by secondary structures of the mRNA. The transcript of the trans-activating element (taR12y) binds to the transcript of the cis-repressive element, hence the RBS is not blocked anymore. The two elements build a riboregulator that decreases leakiness of pLuxR. The expression level of LuxI is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. A biologically neutral spacer sequence was designed using the web application R2oDNA[34]. The p15A is present at a copy number of approximately 15 to 25[35].
Expression of sfGFP and LuxI is induced when LuxR bound to 3OC6-HSL bind to pLuxR. The expression level of sfGFP and LuxI is influenced by the RBS [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]. The p15A is present at a copy number of approximately 15 to 25[35].
 "
"