Team:LMU-Munich/Team/Collaborations
From 2014.igem.org
| (123 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Team:LMU-Munich/Playground/menu}} | {{Template:Team:LMU-Munich/Playground/menu}} | ||
| + | <html> | ||
| + | <section> | ||
| + | <section> | ||
| + | <div class="text-width"> | ||
| + | </html> | ||
| + | = Collaborations = | ||
| + | Collaborations and the exchange of knowledge were important aspects during our iGEM participation. Therefore we had an intensive collaboration with the iGEM team Groningen (Netherlands) and laid a high priority on the exchange with the Rathenau Institute also located in the Netherlands. In addition we also took part in the Interlab Study of this year´s iGEM competition. | ||
| - | = | + | <html> |
| - | + | </div> | |
| + | </section> | ||
| + | <section class="bg-color-2"> | ||
| + | <div> | ||
| + | </html> | ||
== Interlab Study == | == Interlab Study == | ||
=== Background: === | === Background: === | ||
| - | The aim of the Interlab study was to evaluate three devices consisting of different Anderson promoters fused to GFP in two different backbones: | + | The aim of the Interlab study was to evaluate three devices consisting of different Anderson promoters fused to GFP in two different backbones: the high-copy plasmid [http://parts.igem.org/Part:pSB1C3 pSB1C3] (100-300 copies/cell) and the low-to-medium-copy plasmid [http://parts.igem.org/Part:pSB3K3 pSB3K3] (20-30 copies/cell). The devices were distributed by iGEM and the flourescence evaluation method was free of choice. For more details check: [https://2014.igem.org/Tracks/Measurement/Interlab_study Interlab Study@iGEM] |
=== The devices: === | === The devices: === | ||
| - | Device one: BBa_I20260 consisting of J23101-B0032-E0040-B0015 in the backbone | + | *'''Device one:''' BBa_I20260 consisting of J23101-B0032-E0040-B0015 in the backbone pSB3K3 <br> |
| - | Device two: BBa_J23101 + BBa_E0240 each in the backbone | + | :'''Status:''' ready for evaluation |
| - | Device three: BBa_J23115 + BBa_E0240 each in the backbone | + | *'''Device two:''' BBa_J23101 + BBa_E0240, each in the backbone pSB1C3 |
| + | :'''Status:''' cloning before evaluation | ||
| + | *'''Device three:''' BBa_J23115 + BBa_E0240, each in the backbone pSB1C3 | ||
| + | :'''Status:''' cloning before evaluation | ||
| + | |||
=== The cloning: === | === The cloning: === | ||
| - | Devices two and three were built from their individual parts. | + | Devices two and three were built from their individual parts. The plasmids containing the promotors (BBa_J23101 and BBa_J23115) were cut open with SpeI-HF and PstI-HF restriction enzymes. The XbaI and PstI-HF digested GFP (BBa_E0240) was then ligated downstream of the promotors. Both devices were transformed into DH5α, plated out on chloramphenicol [35mg/µl] LB-agar plates and tested via colony-PCR to identify positive clones. For verification PCR-positive clones were sequenced: |
| - | [[Media:LMU14_Interlab_5_091014_BBa_J23115+BBa_E0240.txt | | + | [[Media:LMU14_Interlab_4_091014_BBa_J23101+BBa_E0240.txt | device two]] and |
| + | [[Media:LMU14_Interlab_5_091014_BBa_J23115+BBa_E0240.txt | device three]] | ||
| - | === Flow | + | === Flow cytometry analysis: === |
| - | For | + | For each measurement 50 µl of an overnight culture were first incubated with 1 µl of the membrane stain FM4-64 (final concentration: 4ng/µl) at 37°C for 60 minutes. This is necessary to differentiate between dust or cell particles and intact cells during the measurement. After the incubation, 1 µl of each sample was diluted in 200 µl phosphate buffered saline solution and then evaluated via flow cytometry. In order to generate meaningful results the described measurements were perfomed three times with technical replicates. |
| + | |||
| + | For the analysis, all events were gated for a signal in the red channel (which corresponds to intact cells). For those events, the gfp-fluorescence was measured in fluorescence units. The graph below shows the avarage of the geometric mean of the gfp signals from 3 experiments. The empty DH5α strain was used as control for auto fluorescence of the cells. | ||
| + | |||
| + | For details please check our Interlab study submission: [[Media:LMU14_Interlab_3_091014.pdf | LMU-Munich Interlab form]]<br> | ||
=== Results: === | === Results: === | ||
| - | <br> | + | <html> |
| - | [ | + | <div id="container" class="chart-width" style=" height: 600px;"></div> |
| - | [[ | + | <script> |
| - | < | + | $(function () { |
| + | $('#container').highcharts({ | ||
| + | chart: { | ||
| + | type: 'column', | ||
| + | backgroundColor: null, | ||
| + | plotBorderColor: '#000000', | ||
| + | plotBorderWidth: 2 | ||
| + | }, | ||
| + | color:'#33CC33', | ||
| + | title: { | ||
| + | text: 'GFP fluorescence', | ||
| + | style: { | ||
| + | fontSize: '28px'/*, | ||
| + | fontFamily: 'Verdana, sans-serif',*/ | ||
| + | |||
| + | }, | ||
| + | }, | ||
| + | subtitle: { | ||
| + | text: 'measurement of three technical replicates', | ||
| + | style: { | ||
| + | fontSize: '18px'/*, | ||
| + | fontFamily: 'Verdana, sans-serif',*/ | ||
| + | |||
| + | }, | ||
| + | }, | ||
| + | xAxis: { | ||
| + | type: 'category', | ||
| + | labels: { | ||
| + | rotation: 0, | ||
| + | style: { | ||
| + | fontSize: '18px'/*, | ||
| + | fontFamily: 'Verdana, sans-serif',*/ | ||
| + | } | ||
| + | } | ||
| + | }, | ||
| + | yAxis: { | ||
| + | type: 'logarithmic', | ||
| + | gridLineWidth: 0, | ||
| + | minorGridLineWidth: 0, | ||
| + | min: 1, | ||
| + | title: { | ||
| + | text: 'relativ fluorescence units (millions; geometric mean)', | ||
| + | style: { | ||
| + | fontSize: '18px', | ||
| + | fontFamily: 'Arial, Helvetica, sans-serif', | ||
| + | |||
| + | } | ||
| + | } | ||
| + | }, | ||
| + | credits: false, | ||
| + | legend: { | ||
| + | enabled: false | ||
| + | }, | ||
| + | plotOptions: { | ||
| + | column: { | ||
| + | pointPadding: 0.3, | ||
| + | color: '#33CC33', | ||
| + | borderWidth:2, | ||
| + | borderColor:'#000000' | ||
| + | } | ||
| + | }, | ||
| + | |||
| + | tooltip: { | ||
| + | shared: true, | ||
| + | headerFormat: '<span style="font-size: 13px;font-weight:bold;"> {point.key}</span><br/>', | ||
| + | useHTML: true | ||
| + | }, | ||
| + | |||
| + | |||
| + | series: [{ | ||
| + | name: 'Data', | ||
| + | data: [ | ||
| + | ['DH5α', 203.5], | ||
| + | ['DH5α pSB1C3 K823005-E0240', 17997.7], | ||
| + | ['DH5α pSB1C3 K823012-E0240', 531.3], | ||
| + | ['DH5α pSB3K3 BBa_I20260', 3823.3] | ||
| + | ], | ||
| + | marker: { | ||
| + | enabled: false | ||
| + | }, | ||
| + | tooltip: { | ||
| + | pointFormat: '<span style="font-size:13px">{series.name}</span>: {point.y}<br/>', | ||
| + | valueDecimals: 0 | ||
| + | } | ||
| + | }, { | ||
| + | color: '#FF0000', | ||
| + | name: 'Error range', | ||
| + | type: 'errorbar', | ||
| + | data: [[200,207],[16981.6,19113.7],[448.5,614.2],[3116.3,4530.4]], | ||
| + | tooltip: { | ||
| + | pointFormat: '<span style="font-size:13px">{series.name}</span>: {point.low}-{point.high}' | ||
| + | }, | ||
| + | stemWidth: 5, | ||
| + | whiskerLength: 5, | ||
| + | color: '#000000' | ||
| + | }] | ||
| + | }); | ||
| + | }); | ||
| + | </script> | ||
| + | </html> | ||
| + | <br> | ||
| + | The results indicate that the promoter BBa_K823005 shows the highest expression of GFP, followed by the device BBa_I20260 (BBa_K823005 + B0032). The lowest GFP expression was measured with the device carrying BBa_K823012. <br> | ||
| + | === Discussion === | ||
| + | The results generated here show a 10-fold higher expression read out with GFP for [http://parts.igem.org/Part:BBa_K823005 BBa_K823005] compared to [http://parts.igem.org/Part:BBa_K823012 BBa_K823012]. Having a closer look at the [http://parts.igem.org/Part:BBa_I20260 BBa_I20260] device, the utilization of the low to medium-copy plasmid [http://parts.igem.org/Part:pSB3K3 pSB3K3] explains the lower expression of GFP compared to [http://parts.igem.org/Part:BBa_K823005 BBa_K823005], despite containing the same insert.<br> | ||
| + | === Outlook === | ||
| + | |||
| + | The LMU-iGEM team is looking forward to the presentation of iGEM HQ results, where we hope to see a mixture of different methods utilized - hopefully with similar outcomes. We are curious how the absolute and relative results might differ; this might be a milestone towards ensuring reproducibility of scientific data produced by a huge number of different labs and methods. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </section> | ||
| + | <section class="bg-color-3"> | ||
| + | <div> | ||
| + | </html> | ||
== Collaboration with Team Groningen == | == Collaboration with Team Groningen == | ||
| + | [[File:Groningen_2014_logo.png|thumb|300px|right]] | ||
| + | On August 1st, not only the collaboration with the Rathenau Instituut but also the cooperation with [https://2014.igem.org/Team:Groningen iGEM Team Groningen], was kicked off. During the first group chat with the Rathenau Institute, we were introduced to members of the iGEM Team Groningen that had with their 'Lacto Aid' a quite similar project idea to our BaKillus. As we pursue the same idea but work with different chassis, different targets and aim for different applications, we decided not to see them as rivals but rather use that chance to learn from their experiences and also support them, when help is required. | ||
| + | From then on, we held regular Skype meetings and supported each other by exchanging views and thoughts about the feasibility and social desirability of our applications inspired by the guidelines of the Rathenau scenarios. Encouraged by this first steps, we also helped each other out in the lab. | ||
| + | |||
| + | === Efforts of team Groningen=== | ||
| + | |||
| + | As our BaKillus targets ''Staphylococcus aureus'', but we do not have access to S2 laboratories, and thus unfortunately cannot experiment with this bacterium. Our collaboration partner kindly provided us help by testing the subtilin producer ''Bacillus subtilis'' ATCC6633 in a spot-on-lawn assay on different ''S. aureus'' strains, including MRSA. Fortunately, the results could prove subtilin has inhibitory impact on ''S.aureus'' and thus, serves as excellent killing strategy for our project. | ||
| + | |||
| + | [[File:LMU14 killing subtilin MRSA.png|thumb|700px|center|Fig. 1. Spot-on-lawn assay with the subtilin producer on three different ''S. aureus'' lawns]] | ||
| + | |||
| + | The results are part of our killing subproject and can be seen [https://2014.igem.org/Team:LMU-Munich/Project/Bakillus here] or detailed information downloaded [[Media:LMU14_Groningen_Report_Munchen.pdf|here]]. | ||
| + | |||
| + | === Work done in our Lab === | ||
| + | |||
| + | On Monday 13th of October we received parts to be evaluated in ''Bacillus subtilis'' from the iGEM Team Groningen. | ||
| + | The goal was to evaluate Promoters fused to a super-folded Green Fluorescent Protein (sfGFP) towards their activity in ''[https://2014.igem.org/Team:LMU-Munich/Project/B_subtilis B. subtilis]''. In order to do so we had to bring the BioBricks into our integrative BioBrick compatible backbone [http://parts.igem.org/Part:BBa_J179000 pSB1C]. The final analysis was done using fluorescent-activated cell sorting ([https://static.igem.org/mediawiki/2014/0/02/LMU_Munich14_FACS.pdf FACS]). The results of the strains carrying the Promoter-sfGFP insertion were compared to the ''[https://2014.igem.org/Team:LMU-Munich/Project/B_subtilis B. subtilis]'' W168 strain. | ||
| + | |||
| + | The following Promoters were analyzed: | ||
| + | [http://parts.igem.org/Part:BBa_K1033219 CP1], | ||
| + | [http://parts.igem.org/Part:BBa_K1033221 CP11], | ||
| + | [http://parts.igem.org/Part:BBa_K1033222 CP29], | ||
| + | [http://parts.igem.org/Part:BBa_K1033223 CP30], | ||
| + | [http://parts.igem.org/Part:BBa_K1033224 CP41], | ||
| + | [http://parts.igem.org/Part:BBa_K1033225 CP44] | ||
| + | |||
| + | The data achieved via FACS shows no change in distribution of FL1-A intensity. This strongly inidcates, that the promoter-''sfGFP'' constructs are not active when integrated into the ''[https://2014.igem.org/Team:LMU-Munich/Project/B_subtilis B. subtilis]'' chromosome. This might however be not only due to an lack of Promoter activity. To further invesitgate a quantitative assay using promoter-''lux'' fusions should be carried out. | ||
| + | |||
| + | <html> | ||
| + | <div class="box-center"> | ||
| + | <ul class="bxgallery"> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/e/ee/LMU14_Collaboration_CP1.PNG" alt="CP1 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/a/ae/LMU14_Collaboration_CP11.PNG" alt="CP11 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/d/d3/LMU14_Collaboration_CP29.PNG" alt="CP29 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/4/42/LMU14_Collaboration_CP30.PNG" alt="CP30 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/6/64/LMU14_Collaboration_CP41.PNG" alt="CP41 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | <li><img src="https://static.igem.org/mediawiki/2014/e/e7/LMU14_Collaboration_CP44.PNG" alt="CP44 (red, triplicates) and Wildtype (black, duplicates)"/></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <i>Table 1: FACS Data</i> | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !<b>Promoter</b> | ||
| + | !<b>Count</b> | ||
| + | !<b>Mean FL1-A</b> | ||
| + | |- | ||
| + | !rowspan="1"|<b>W168 (Wild type strain)</b> | ||
| + | |627609 | ||
| + | |155.95 | ||
| + | |- | ||
| + | !rowspan="1"|<b>W168 (Wild type strain)</b> | ||
| + | |86172 | ||
| + | |186.03 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP1</b> | ||
| + | |679126 | ||
| + | |163.13 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP1</b> | ||
| + | |587189 | ||
| + | |160.54 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP1</b> | ||
| + | |644350 | ||
| + | |161.32 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP11</b> | ||
| + | |558153 | ||
| + | |164.57 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP11</b> | ||
| + | |371964 | ||
| + | |194.71 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP11</b> | ||
| + | |169210 | ||
| + | |188.28 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP29</b> | ||
| + | |50758 | ||
| + | |310.50 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP29</b> | ||
| + | |399988 | ||
| + | |173.85 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP29</b> | ||
| + | |464802 | ||
| + | |158.56 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP30</b> | ||
| + | |665875 | ||
| + | |155.95 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP30</b> | ||
| + | |657525 | ||
| + | |155.84 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP30</b> | ||
| + | |463322 | ||
| + | |157.94 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP41</b> | ||
| + | |463478 | ||
| + | |157.61 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP41</b> | ||
| + | |463578 | ||
| + | |157.95 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP41</b> | ||
| + | |463237 | ||
| + | |159.17 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP44</b> | ||
| + | |463306 | ||
| + | |158.39 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP44</b> | ||
| + | |463375 | ||
| + | |157.43 | ||
| + | |- | ||
| + | !rowspan="1"|<b>CP44</b> | ||
| + | |463508 | ||
| + | |157.34 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </section> | ||
| + | <section class="bg-color-1"> | ||
| + | <div> | ||
| + | </html> | ||
| + | |||
| + | == A proposal of the German iGEM Teams concerning Intellectual Property == | ||
| + | |||
| + | During the meetup of the German iGEM teams from 23rd to 25th May a [https://2014.igem.org/Meetups:May_LMU-Munich/Workshops workshop] also took place in which amongst others we discussed the topic of bioethics. Moral questions were addressed, regarding the value of life and human influence on it, as well as questions dealing with the possible socioeconomic effects of synthetic biology. | ||
| + | |||
| + | Especially the topic of an open source vs. patent controlled field accounted for a large part of the discussion. During the discussion one student brought up the point that the legal status of parts in the parts registry remains unclear and that there are parts (e.g. [http://parts.igem.org/Part:BBa_K180009 BBa_K180009]) where it only becomes clear upon a closer that the rights are company–owned. This issue (that the legal status of parts in the registry remains uncertain) is also mentioned in a recent article published by Nature [1]: | ||
| + | |||
| + | ''"[N]o one can say with any certainty how many of these parts are themselves entirely free of patent claims."'' | ||
| + | |||
| + | We, the German iGEM teams, therefore like to suggest the addition of a new feature to the parts registry: | ||
| + | |||
| + | A dedicated data field of license information for each BioBrick part. | ||
| + | |||
| + | For the implementation, we propose to introduce two new fields to the BioBrick part entries in the registry: | ||
| + | :#A string property "LicenseInfo" | ||
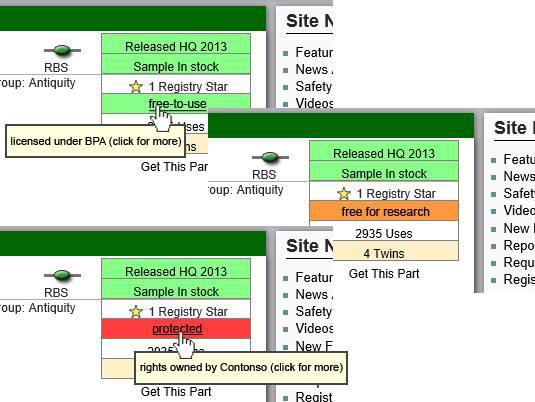
| + | :#A traffic light property (grey, green, yellow, red) to indicate the level of legal protection (unknown, BPA-like, free for research purposes, heavily protected) | ||
| + | |||
| + | |||
| + | [[File:TU-BS_Part_licence_info.png|center]] | ||
| + | |||
| + | |||
| + | Implementing this feature would in our opinion further clarify and extend the parts info, provide a machine-readable format and thus improve future entries. With the emerging Entrepreneurship track and applications getting closer to industrial realization, the legal status becomes more and more relevant. It would also raise awareness to the topic of the legal status of parts. This could lead to a debate which could further promote the idea of open source, ideally. At the same time we hope that the examination of most parts will show that they are indeed free of restrictive legal protections. | ||
| + | |||
| + | :The German iGEM Teams, | ||
| + | |||
| + | [https://2014.igem.org/Team:Aachen Aachen] | ||
| + | [https://2014.igem.org/Team:Berlin Berlin] | ||
| + | [https://2014.igem.org/Team:Bielefeld-CeBiTec Bielefeld-CeBiTec] | ||
| + | [https://2014.igem.org/Team:Braunschweig Braunschweig] | ||
| + | [https://2014.igem.org/Team:Freiburg Freiburg] | ||
| + | [https://2014.igem.org/Team:Goettingen Goettingen] | ||
| + | [https://2014.igem.org/Team:Hannover Hannover] | ||
| + | [https://2014.igem.org/Team:Heidelberg Heidelberg] | ||
| + | [https://2014.igem.org/Team:LMU-Munich LMU-Munich] | ||
| + | [https://2014.igem.org/Team:Marburg Marburg] | ||
| + | [https://2014.igem.org/Team:Saarland Saarland] | ||
| + | [https://2014.igem.org/Team:Tuebingen Tuebingen] | ||
| + | [https://2014.igem.org/Team:TU_Darmstadt TU Darmstadt] | ||
| + | |||
| + | References: | ||
| + | *[http://www.nature.com/news/synthetic-biology-cultural-divide-1.15149| Bryn Nelson ‘Synthetic Biology: Cultural Divide ’, Nature 509, 152–154, 08 May 2014] | ||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </section> | ||
| + | <section class="bg-color-2"> | ||
| + | <div> | ||
| + | </html> | ||
| + | |||
| + | == Collaboration with Team Virginia == | ||
| + | |||
| + | <html> | ||
| + | <head> | ||
| + | <style> | ||
| + | #virginiabadge{ | ||
| + | background: url("https://static.igem.org/mediawiki/2014/6/6a/Virginia_Badge2.png"); | ||
| + | background-size:250px; | ||
| + | width: 250px; | ||
| + | height: 185px; | ||
| + | background-repeat:no-repeat; | ||
| + | } | ||
| + | |||
| + | #virginiabadge:hover{ | ||
| + | background: url("https://static.igem.org/mediawiki/2014/2/2b/File-Virginia-Images-Badge1.png"); | ||
| + | background-repeat:no-repeat; | ||
| + | width: 250px; | ||
| + | height: 185px; | ||
| + | background-size:250px; | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | |||
| + | <body> | ||
| + | <div id="virginiabadge"> | ||
| + | <a href= | ||
| + | "https://2014.igem.org/Team:Virginia/HumanPractices"><img height="180" | ||
| + | src="https://static.igem.org/mediawiki/2014/b/b0/Virginia_Transparent.png" | ||
| + | width="250"></a> | ||
| + | </div> | ||
| + | </body> | ||
| + | The global synthetic biology awareness and acceptance survey designed by the Virginia iGEM-team was a great opportunity to generate a lot of data, <br>since more than 40 teams participated! | ||
| + | </html> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | </div> | ||
| + | </section> | ||
| + | </section> | ||
| + | </html> | ||
{{Template:Team:LMU-Munich/Playground/footer}} | {{Template:Team:LMU-Munich/Playground/footer}} | ||
<html><script>initiateNavigation("team");</script></html> | <html><script>initiateNavigation("team");</script></html> | ||
Latest revision as of 03:23, 18 October 2014
Collaborations
Collaborations and the exchange of knowledge were important aspects during our iGEM participation. Therefore we had an intensive collaboration with the iGEM team Groningen (Netherlands) and laid a high priority on the exchange with the Rathenau Institute also located in the Netherlands. In addition we also took part in the Interlab Study of this year´s iGEM competition.
Interlab Study
Background:
The aim of the Interlab study was to evaluate three devices consisting of different Anderson promoters fused to GFP in two different backbones: the high-copy plasmid [http://parts.igem.org/Part:pSB1C3 pSB1C3] (100-300 copies/cell) and the low-to-medium-copy plasmid [http://parts.igem.org/Part:pSB3K3 pSB3K3] (20-30 copies/cell). The devices were distributed by iGEM and the flourescence evaluation method was free of choice. For more details check: Interlab Study@iGEM
The devices:
- Device one: BBa_I20260 consisting of J23101-B0032-E0040-B0015 in the backbone pSB3K3
- Status: ready for evaluation
- Device two: BBa_J23101 + BBa_E0240, each in the backbone pSB1C3
- Status: cloning before evaluation
- Device three: BBa_J23115 + BBa_E0240, each in the backbone pSB1C3
- Status: cloning before evaluation
The cloning:
Devices two and three were built from their individual parts. The plasmids containing the promotors (BBa_J23101 and BBa_J23115) were cut open with SpeI-HF and PstI-HF restriction enzymes. The XbaI and PstI-HF digested GFP (BBa_E0240) was then ligated downstream of the promotors. Both devices were transformed into DH5α, plated out on chloramphenicol [35mg/µl] LB-agar plates and tested via colony-PCR to identify positive clones. For verification PCR-positive clones were sequenced: device two and device three
Flow cytometry analysis:
For each measurement 50 µl of an overnight culture were first incubated with 1 µl of the membrane stain FM4-64 (final concentration: 4ng/µl) at 37°C for 60 minutes. This is necessary to differentiate between dust or cell particles and intact cells during the measurement. After the incubation, 1 µl of each sample was diluted in 200 µl phosphate buffered saline solution and then evaluated via flow cytometry. In order to generate meaningful results the described measurements were perfomed three times with technical replicates.
For the analysis, all events were gated for a signal in the red channel (which corresponds to intact cells). For those events, the gfp-fluorescence was measured in fluorescence units. The graph below shows the avarage of the geometric mean of the gfp signals from 3 experiments. The empty DH5α strain was used as control for auto fluorescence of the cells.
For details please check our Interlab study submission: LMU-Munich Interlab form
Results:
The results indicate that the promoter BBa_K823005 shows the highest expression of GFP, followed by the device BBa_I20260 (BBa_K823005 + B0032). The lowest GFP expression was measured with the device carrying BBa_K823012.
Discussion
The results generated here show a 10-fold higher expression read out with GFP for [http://parts.igem.org/Part:BBa_K823005 BBa_K823005] compared to [http://parts.igem.org/Part:BBa_K823012 BBa_K823012]. Having a closer look at the [http://parts.igem.org/Part:BBa_I20260 BBa_I20260] device, the utilization of the low to medium-copy plasmid [http://parts.igem.org/Part:pSB3K3 pSB3K3] explains the lower expression of GFP compared to [http://parts.igem.org/Part:BBa_K823005 BBa_K823005], despite containing the same insert.
Outlook
The LMU-iGEM team is looking forward to the presentation of iGEM HQ results, where we hope to see a mixture of different methods utilized - hopefully with similar outcomes. We are curious how the absolute and relative results might differ; this might be a milestone towards ensuring reproducibility of scientific data produced by a huge number of different labs and methods.
Collaboration with Team Groningen
On August 1st, not only the collaboration with the Rathenau Instituut but also the cooperation with iGEM Team Groningen, was kicked off. During the first group chat with the Rathenau Institute, we were introduced to members of the iGEM Team Groningen that had with their 'Lacto Aid' a quite similar project idea to our BaKillus. As we pursue the same idea but work with different chassis, different targets and aim for different applications, we decided not to see them as rivals but rather use that chance to learn from their experiences and also support them, when help is required. From then on, we held regular Skype meetings and supported each other by exchanging views and thoughts about the feasibility and social desirability of our applications inspired by the guidelines of the Rathenau scenarios. Encouraged by this first steps, we also helped each other out in the lab.
Efforts of team Groningen
As our BaKillus targets Staphylococcus aureus, but we do not have access to S2 laboratories, and thus unfortunately cannot experiment with this bacterium. Our collaboration partner kindly provided us help by testing the subtilin producer Bacillus subtilis ATCC6633 in a spot-on-lawn assay on different S. aureus strains, including MRSA. Fortunately, the results could prove subtilin has inhibitory impact on S.aureus and thus, serves as excellent killing strategy for our project.
The results are part of our killing subproject and can be seen here or detailed information downloaded here.
Work done in our Lab
On Monday 13th of October we received parts to be evaluated in Bacillus subtilis from the iGEM Team Groningen. The goal was to evaluate Promoters fused to a super-folded Green Fluorescent Protein (sfGFP) towards their activity in B. subtilis. In order to do so we had to bring the BioBricks into our integrative BioBrick compatible backbone [http://parts.igem.org/Part:BBa_J179000 pSB1C]. The final analysis was done using fluorescent-activated cell sorting (FACS). The results of the strains carrying the Promoter-sfGFP insertion were compared to the B. subtilis W168 strain.
The following Promoters were analyzed: [http://parts.igem.org/Part:BBa_K1033219 CP1], [http://parts.igem.org/Part:BBa_K1033221 CP11], [http://parts.igem.org/Part:BBa_K1033222 CP29], [http://parts.igem.org/Part:BBa_K1033223 CP30], [http://parts.igem.org/Part:BBa_K1033224 CP41], [http://parts.igem.org/Part:BBa_K1033225 CP44]
The data achieved via FACS shows no change in distribution of FL1-A intensity. This strongly inidcates, that the promoter-sfGFP constructs are not active when integrated into the B. subtilis chromosome. This might however be not only due to an lack of Promoter activity. To further invesitgate a quantitative assay using promoter-lux fusions should be carried out.
Table 1: FACS Data
| Promoter | Count | Mean FL1-A |
|---|---|---|
| W168 (Wild type strain) | 627609 | 155.95 |
| W168 (Wild type strain) | 86172 | 186.03 |
| CP1 | 679126 | 163.13 |
| CP1 | 587189 | 160.54 |
| CP1 | 644350 | 161.32 |
| CP11 | 558153 | 164.57 |
| CP11 | 371964 | 194.71 |
| CP11 | 169210 | 188.28 |
| CP29 | 50758 | 310.50 |
| CP29 | 399988 | 173.85 |
| CP29 | 464802 | 158.56 |
| CP30 | 665875 | 155.95 |
| CP30 | 657525 | 155.84 |
| CP30 | 463322 | 157.94 |
| CP41 | 463478 | 157.61 |
| CP41 | 463578 | 157.95 |
| CP41 | 463237 | 159.17 |
| CP44 | 463306 | 158.39 |
| CP44 | 463375 | 157.43 |
| CP44 | 463508 | 157.34 |
A proposal of the German iGEM Teams concerning Intellectual Property
During the meetup of the German iGEM teams from 23rd to 25th May a workshop also took place in which amongst others we discussed the topic of bioethics. Moral questions were addressed, regarding the value of life and human influence on it, as well as questions dealing with the possible socioeconomic effects of synthetic biology.
Especially the topic of an open source vs. patent controlled field accounted for a large part of the discussion. During the discussion one student brought up the point that the legal status of parts in the parts registry remains unclear and that there are parts (e.g. [http://parts.igem.org/Part:BBa_K180009 BBa_K180009]) where it only becomes clear upon a closer that the rights are company–owned. This issue (that the legal status of parts in the registry remains uncertain) is also mentioned in a recent article published by Nature [1]:
"[N]o one can say with any certainty how many of these parts are themselves entirely free of patent claims."
We, the German iGEM teams, therefore like to suggest the addition of a new feature to the parts registry:
A dedicated data field of license information for each BioBrick part.
For the implementation, we propose to introduce two new fields to the BioBrick part entries in the registry:
- A string property "LicenseInfo"
- A traffic light property (grey, green, yellow, red) to indicate the level of legal protection (unknown, BPA-like, free for research purposes, heavily protected)
Implementing this feature would in our opinion further clarify and extend the parts info, provide a machine-readable format and thus improve future entries. With the emerging Entrepreneurship track and applications getting closer to industrial realization, the legal status becomes more and more relevant. It would also raise awareness to the topic of the legal status of parts. This could lead to a debate which could further promote the idea of open source, ideally. At the same time we hope that the examination of most parts will show that they are indeed free of restrictive legal protections.
- The German iGEM Teams,
Aachen Berlin Bielefeld-CeBiTec Braunschweig Freiburg Goettingen Hannover Heidelberg LMU-Munich Marburg Saarland Tuebingen TU Darmstadt
References:
- [http://www.nature.com/news/synthetic-biology-cultural-divide-1.15149| Bryn Nelson ‘Synthetic Biology: Cultural Divide ’, Nature 509, 152–154, 08 May 2014]

Hi there!
Welcome to our Wiki! I'm BaKillus, the pathogen-hunting microbe, and I'll guide you on this tour through our project. If you want to learn more about a specific step, you can simply close the tour and come back to it anytime you like. So let's start!

What's the problem?
First of all, what am I doing here? The problem is, pathogenic bacteria all around the world are becoming more and more resistant against antimicrobial drugs. One major reason for the trend is the inappropriate use of drugs. With my BaKillus super powers, I want to reduce this misuse and thus do my part to save global health.

Sensing of pathogens
To combat the pathogenic bacteria, I simply eavesdrop on their communication. Bacteria talk with each other via quorum sensing systems, which I use to detect them and trigger my responses.

Adhesion
The more specific and effective I can use my powers, the lower the danger is of provoking new resistance development. So I catch pathogens whenever I get hold of them and stick to them until my work is done.

Killing
Talking about my work - killing pathogens is finally what I am made for. In response to quorum sensing molecules of the pathogens, I export a range of antimicrobial substances leading to dissipation of biofilms and the killing of the targeted bacteria.

Suicide switch
When the job is done and all the bad guys are finished, you don't need a super hero anymore. So after fulfilling my work I say goodbye to the world by activating my suicide switch.

Application
Of course I'm not only a fictional hero, but a very real one. In two different prototypes, I could be used for diagnosis or treatment of pathogen-caused diseases. However, there is still a whole lot of regulational and economical questions that have to be answered before.

See you!
So now you know my short story - and it is time for me to return to my fight for a safer world. Feel free to take a closer look on my super powers, the process of my development or the plans for a medical application.
 "
"