Team:Aachen/Project/Gal3
From 2014.igem.org
(→Achievements) |
|||
| Line 71: | Line 71: | ||
<span class="anchor" id="alternativesensing"></span> | <span class="anchor" id="alternativesensing"></span> | ||
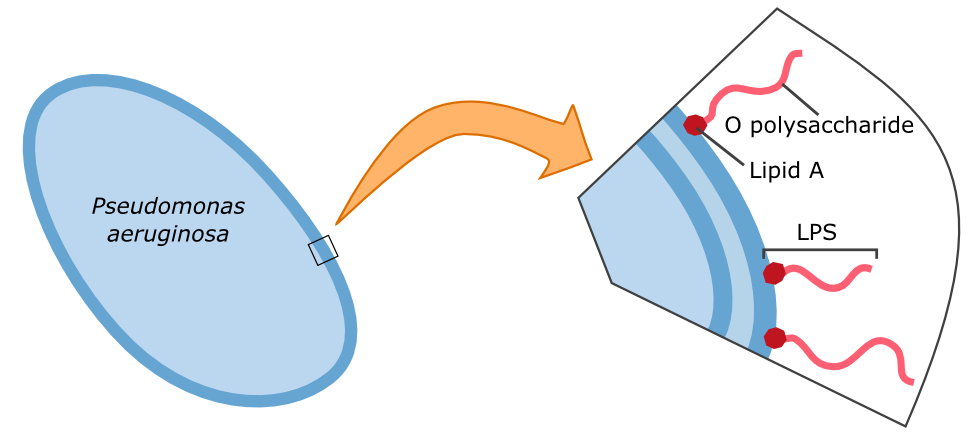
| - | {{Team:Aachen/FigureFloat|Aachen_14-10-09_Pseudomonas_LPS_iNB.png|title=Cell wall composition of ''P. aeruginosa''|subtitle=Gram-negative bacteria have two cell membranes. The LPS are embedded in the outer membrane and are composed of a lipid and an O polysaccharide.|width=420px}} | + | {{Team:Aachen/FigureFloat|Aachen_14-10-09_Pseudomonas_LPS_iNB.png|title=Cell wall composition of ''P. aeruginosa''|subtitle=Gram-negative bacteria have two cell membranes. The LPS are embedded in the outer membrane and are composed of a lipid and an O polysaccharide.|width=420px}} |
| - | Characteristic parts of the '''lipopolysaccharide structure (LPS)''' of ''P. aeruginosa'' can be bound by galectin-3. Specifically, the O polysaccharide (see figure on the left) of the LPS is recognized by galectin-3. Therefore, this specific binding of galectin-3 enables the construction of a fluorescent based detection system. A fusion protein of galectin-3 and a reporter protein, such as a fluorescent protein, can be built and applied in the detection of ''P. aeruginosa''. | + | Characteristic parts of the '''lipopolysaccharide structure (LPS)''' of ''P. aeruginosa'' can be bound by galectin-3. Specifically, the O polysaccharide (see figure on the left) of the LPS is recognized by galectin-3. Therefore, this specific binding of galectin-3 enables the construction of a fluorescent based detection system. A fusion protein of galectin-3 and a reporter protein, such as a fluorescent protein, can be built and applied in the detection of ''P. aeruginosa''. |
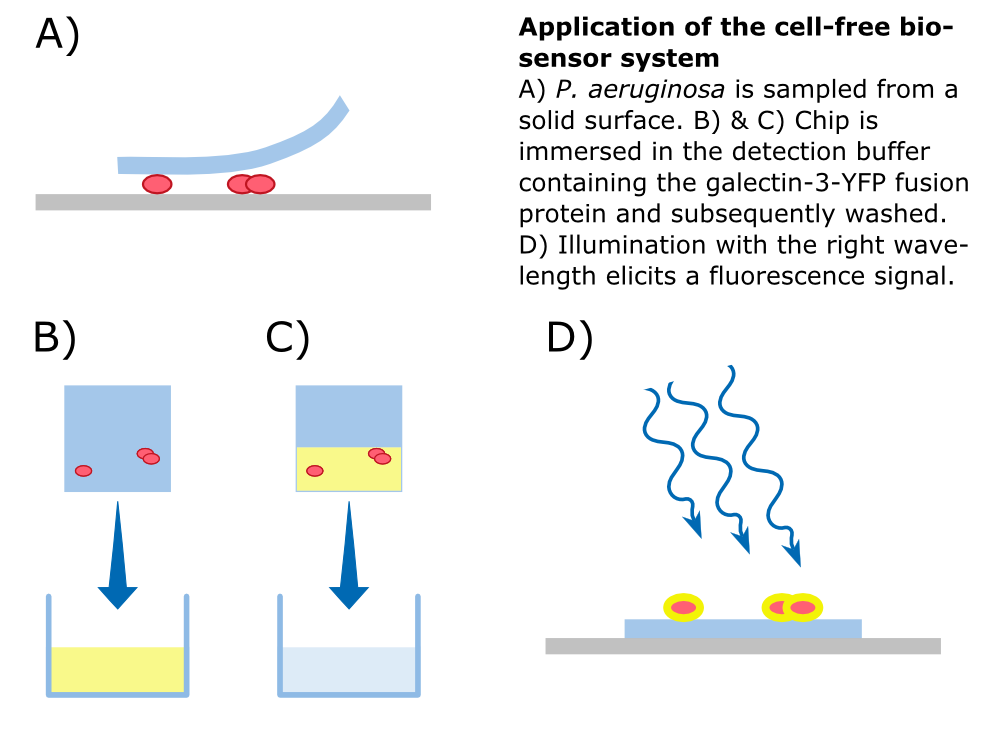
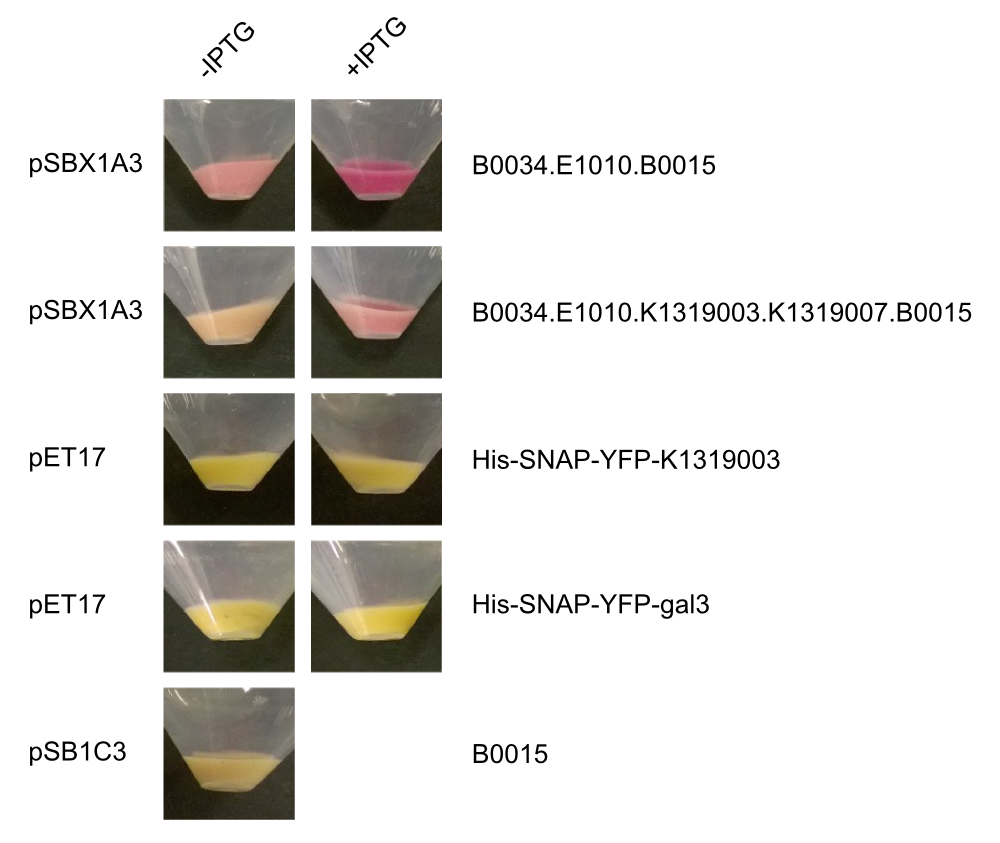
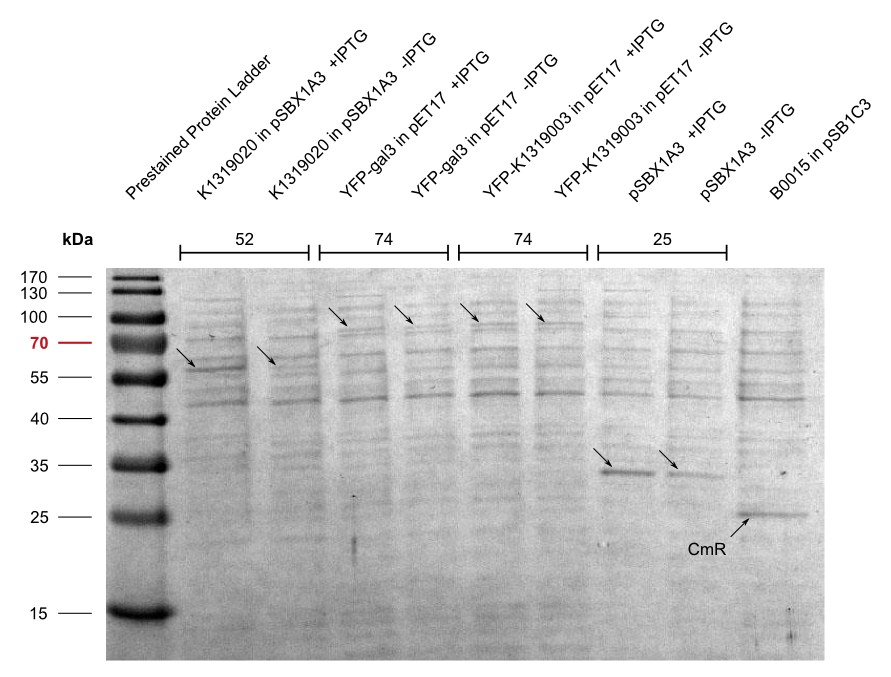
In our approach, a '''galectin-3-YFP fusion protein''' is built and expressed in ''E. coli'', including a his-tag and a snap-tag for purification. The fusion protein can then be incorporated into a '''cell-free biosensor system'''. These biosensors have many advantages over systems that use living cells such as an uncomplicated storage. Furthermore, from a [https://2014.igem.org/Team:Aachen/Safety biosafety] and social acceptance point of view, it is advantageous if the sensor system does not contain alive genetically modified organisms. | In our approach, a '''galectin-3-YFP fusion protein''' is built and expressed in ''E. coli'', including a his-tag and a snap-tag for purification. The fusion protein can then be incorporated into a '''cell-free biosensor system'''. These biosensors have many advantages over systems that use living cells such as an uncomplicated storage. Furthermore, from a [https://2014.igem.org/Team:Aachen/Safety biosafety] and social acceptance point of view, it is advantageous if the sensor system does not contain alive genetically modified organisms. | ||
| Line 94: | Line 94: | ||
With the help of David Schönauer and Alan Mertens from the RWTH Aachen Institute of Biotechnolgy we then purified the fusion protein using FPLC. | With the help of David Schönauer and Alan Mertens from the RWTH Aachen Institute of Biotechnolgy we then purified the fusion protein using FPLC. | ||
| - | Subsequently, we aimed to test the binding of the Gal-3 fusion protein to the LPS of ''P. aeruginosa'' as shown previously (Kupper, Böcker, Liu et al., 2013). Apparently, because of insufficient sensitivity of the used fluorescence microscope, this could not be confirmed and would require further experiments, idealy using other detection methods. | + | Subsequently, we aimed to test the binding of the Gal-3 fusion protein to the LPS of ''P. aeruginosa'' as shown previously (Kupper, Böcker, Liu et al., 2013). Apparently, because of insufficient sensitivity of the used fluorescence microscope, this could not be confirmed and would require further experiments, idealy using other detection methods. |
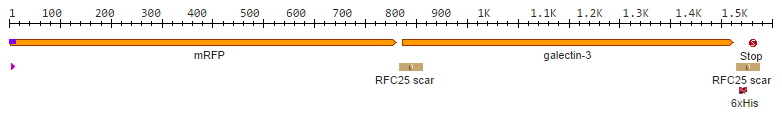
After we received the collection of [https://2014.igem.org/Team:Aachen/Collaborations/Heidelberg pSBX-expression vectors] from Team Heidelberg, we used Gibson assembly to make K1319020 from K1319003 and pSBX1A3, which is the translational unit for an mRFP-Gal3 fusion protein with a C-terminal 6xHis tag: | After we received the collection of [https://2014.igem.org/Team:Aachen/Collaborations/Heidelberg pSBX-expression vectors] from Team Heidelberg, we used Gibson assembly to make K1319020 from K1319003 and pSBX1A3, which is the translational unit for an mRFP-Gal3 fusion protein with a C-terminal 6xHis tag: | ||
Latest revision as of 03:28, 18 October 2014
|
|
|
|
|
 "
"