Team:Freiburg/Content/Results/Light system
From 2014.igem.org
| (46 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | |||
| - | < | + | <div class="row category-row"> |
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: left"> | ||
| + | <div style="position: relative; float: right; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results/Receptor">Go back to The Receptor</div> | ||
| + | <div style="position: relative; float: left;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/4/44/Freiburg2014_Navigation_Arrow_rv.png"> <!-- Pfeil rv--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: right"> | ||
| + | <div style="position: relative; float: left; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results/The_combination">See the Results of the Combination of our Systems</div> | ||
| + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <section id="Light-system"> | ||
| + | |||
| + | <h1>The Light System</h1> | ||
| - | + | <p> Light inducible systems allow spatial and temporal control over gene expression. | |
| - | + | Our goal is the expression of the mCAT-1 receptor through a light inducible | |
| + | system to allow spatiotemporal control of the virus entry into the target cells. | ||
| + | We used two different systems: expression is induced by light of the blue and | ||
| + | red wavelength range. To test the performance of the light systems, we used an | ||
| + | assay where the cells express secreted embryonic alkaline phosphatase (SEAP). | ||
| + | The amount of produced SEAP can be measured by its enzymatic activity. It | ||
| + | breaks down the substrate para-nitrophenylphosphate to the yellow para-nitrophenol, | ||
| + | which can be measured with a spectrophotometer. | ||
| + | </p> | ||
| - | + | <h2 id="Results-LightSystem-Function">Function of the Light System</h2> | |
| - | + | <div class="row category-row"> | |
| - | + | <div class="col-sm-6"> | |
| - | + | <p> Because the proper function of the light system is important for our project, | |
| - | + | we tested two different light systems. The red light system is based on the protein | |
| + | phytochrome B (PhyB) from Arabidopsis thaliana and is activated by light around 660 nm, | ||
| + | whereas the blue light system contains a protein from Avena sativa (AsLOV2) which | ||
| + | requires 465 nm light for activation. | ||
| + | </p> | ||
| + | <p> To test the function of the red light system, we used Chinese hamster ovary cells (CHO) | ||
| + | that are known to work better for this purpose than other cell lines. For the blue | ||
| + | light system, we tested CHO cells and human embryonic kidney cells (HEK-293T). | ||
| + | As reporter the gene seap was transfected together with the genes required for | ||
| + | the implementation of the blue and red light inducible systems. | ||
| + | A SEAP-assay was performed after illumination. Measuring SEAP concentrations in the | ||
| + | supernatant lead us two two important conclusions: First, the blue light system works | ||
| + | more efficient than the red light system; second, the blue light system works better | ||
| + | with HEK-293T cells than with CHO cells (Fig. 1). | ||
| + | </p> | ||
| + | <p> | ||
| + | The plasmids required for the implementation of the light inducible SEAP expression are: | ||
| + | </p> | ||
| + | Red light system: | ||
| + | <ul> | ||
| + | <li>pKM022: SV40_PhyB-VP16_IRES_TetR-PIF16<br/ > constitutive expression of PhyB-VP16 and TetR-PIF6</li> | ||
| + | <li>pKM006: tetO-CMVmin_SEAP<br/ > SEAP under control of the tet operon</li> | ||
| + | </ul> | ||
| + | Blue light system: | ||
| + | <ul> | ||
| + | <li>pKM292: SV40_Gal4BD-LOV<br/ > constitutive expression of Gal4BD-LOV</li> | ||
| + | <li>pKM297: SV40_ePDZ-VP16<br/ > constitutive expression of ePDZ-VP16</li> | ||
| + | <li>pKM084: Gal4UAS-CMVmin_SEAP<br/ > SEAP under control of the Gal4 upstream activating sequence</li> | ||
| + | </ul> | ||
| + | <p> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Project/The_light_system"> | ||
| + | For more information about the light systems click here! | ||
| + | </a> | ||
| + | </p> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | </div> | |
| - | + | <div class="col-sm-6"> | |
| - | + | <figure> | |
| - | + | <a href="https://static.igem.org/mediawiki/2014/e/ef/Freiburg2014_results_light_systems_compared.png"> | |
| - | </figure> | + | <img src="https://static.igem.org/mediawiki/2014/e/ef/Freiburg2014_results_light_systems_compared.png"> |
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header"> | ||
| + | Figure 1: Comparison of the blue light system in CHO cells and in HEK-293T cells and | ||
| + | comparison to the red light system in CHO cells. | ||
| + | </p> | ||
| + | <p class="desc"> | ||
| + | To test the efficiency of the light system, SEAP expression after illumination was determined. | ||
| + | The SEAP assay was performed 24 hours after light exposure. | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Red_light">Labjournal</a> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </div> | ||
| - | + | <div class="row category-row"> | |
| - | + | <div class="col-sm-6"> | |
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/4/4f/Freiburg2014_results_blue_light_system_different_illuminations_seap.png"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/4f/Freiburg2014_results_blue_light_system_different_illuminations_seap.png"> | ||
| + | </a> | ||
| + | <figcaption> | ||
| + | <p class="header"> | ||
| + | Figure 2: Efficiency of the blue light system using different time intervals of illumination. | ||
| + | </p> | ||
| + | <p class="desc"> | ||
| + | To test the efficiency of the light system, SEAP expression after illumination was determined. | ||
| + | The SEAP assay was performed 24 hours after light exposure. Cells were incubated with | ||
| + | blue light for 1 hour, 2.5 hours and 5 hours. | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_Blue_light">Labjournal</a> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <p> | ||
| + | The blue light system is effective in activating the reporter SEAP which was introduced | ||
| + | into the cells before illumination by transfections. As we know | ||
| + | that the duration of illuminating the system with blue light (452 nm) is critical | ||
| + | for its efficiency, we tested various time intervals for illumination. Our results | ||
| + | indicate that five hours of illumination lead to the highest level of SEAP expression (Fig. 2). | ||
| + | </p> | ||
| + | <p> | ||
| + | Another important finding was the specificity of the blue light system. | ||
| + | In the dark controls, we found almost no activation of the SEAP reporter, | ||
| + | leading to a very low background level in our system. | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <h2 id="Results-LightSystem-SEAP-as-Reporter">SEAP as a Reporter</h2> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p> | |
| - | + | In order to test the capability of the blue light system for pattern generation, we transfected HEK-293T cells with the appropriate plasmids (PKM292 and PKM297) using seap as a reporter gene (PKM084). Cells were transfected in suspension and seeded on 96 well plates afterwards. Single wells of these plates were covered with a photo mask that prevents exposure to light. All other wells on the plate were illuminated to induce SEAP expression. SEAP that was secreted into the culture medium leads to a colour change from red to yellow after addition of pNPP (chromogenic substrate) by its phosphorylation. | |
| - | + | </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | <p> |
| + | First we generated a coarse pattern realizing that scattered light also reached wells that were close to the illuminated ones, leading to an unclear pattern. However, the rough shape of the desired pattern is discernible (Fig. 3A, B). | ||
| + | </p> | ||
| - | + | <p> | |
| - | + | Since it was essential to completely prevent light exposure on covered plates, we tested 96 well plates with optically separated wells. We could generate patterns with sharp contours (Fig. 3C, D). | |
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | The colour change of pNPP in cell culture medium could be measured at 405 nm in a plate reader. Setting a distinct threshold made pattern recognition easier. | ||
| + | </p> | ||
| - | + | <figure class="fig-full-width"> | |
| - | + | <a href="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> | |
| - | + | <img class="img-no-pad" src="https://static.igem.org/mediawiki/2014/3/31/Freiburg2014_Results_light_QR_96.jpg"> | |
| - | + | </a> | |
| - | + | <figcaption> | |
| - | + | <p class="header"> | |
| - | + | Figure 3: Generation of patterns on 96-well plates. | |
| - | + | </p> | |
| - | + | <p class="desc"> | |
| - | + | HEK-293T cells were transfected with the blue light system (PKM292, PKM297) and | |
| - | + | light induced SEAP as reporter (PKM084). Single wells were covered with a photo | |
| - | + | mask to prevent cell from light exposure. 24 hours after illumination with blue | |
| - | + | light, a SEAP assay was performed, leading to a colour change of wells containing SEAP. | |
| - | + | (A) Photo mask in heart form, (B) pseudocoloring done at a computer, | |
| + | (C) the same experiment was performed with optically separated wells, | ||
| + | (D) the contrast was increased and the colours of the wells were changed | ||
| + | at a computer. All wells together are forming the word: IGEM. | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_QR_IGEM">Labjournal 1</a> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_QR_Herz">Labjournal 2</a> | ||
| + | </p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ||
| - | + | <p> | |
| + | For generating QR codes with our method 384-well plates were used. Since we had a collaboration | ||
| + | with the iGEM Team Aachen 2014, they made a photo mask for a QR code for us fitting on these plates (Fig. 4). | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Team/Collaboration#Team-Collaboration-Aachen">Collaborations</a>. | ||
| + | </p> | ||
| - | + | <figure class="fig-full-width"> | |
| - | + | <a href="https://static.igem.org/mediawiki/2014/b/b4/Freiburg2014_Results_light_QR_384.jpg"> | |
| - | + | <img class="img-no-pad" src="https://static.igem.org/mediawiki/2014/b/b4/Freiburg2014_Results_light_QR_384.jpg"> | |
| - | + | </a> | |
| - | + | <figcaption> | |
| - | + | <p class="header"> | |
| - | + | Figure 4: Generation of a QR code on a 384 well plate. | |
| - | + | </p> | |
| - | + | <p class="desc"> | |
| - | + | HEK-293T cells were transfected with the blue light system (PKM292, PKM297) | |
| - | + | and light induced SEAP as reporter (PKM084). Single wells were covered with a photo mask | |
| - | + | to prevent cell from light exposure using a photo mask for a QR code (made by iGEM Team Aachen 2014). | |
| - | + | 24 hours after illumination with blue light a SEAP assay was performed leading to colour | |
| - | + | changing of wells containing SEAP. Only a part of the plate was used. (A) Photo mask | |
| - | + | for QR code generation, (B) Picture of the 384-well plate after the SEAP assay, (C) the | |
| - | + | same plate with contrast of the colours increased at a computer. | |
| - | + | <a href="https://2014.igem.org/Team:Freiburg/Notebook/Labjournal#MirjaHarms_QR_384">Labjournal</a> | |
| - | + | </p> | |
| - | + | </figcaption> | |
| - | + | </figure> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | </section> | ||
| + | <div class="row category-row"> | ||
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: left"> | ||
| + | <div style="position: relative; float: right; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results/Receptor">Go back to The Receptor</div> | ||
| + | <div style="position: relative; float: left;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/4/44/Freiburg2014_Navigation_Arrow_rv.png"> <!-- Pfeil rv--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <div class="col-sm-6"> | ||
| + | <div class="container-fluid" style="float: right"> | ||
| + | <div style="position: relative; float: left; margin-top: 4px;"> | ||
| + | <a href="https://2014.igem.org/Team:Freiburg/Results/The_combination">See the Results of the Combination of our Systems</div> | ||
| + | <div style="position: relative; float: right;"> <img class="img-no-border" style="max-width: 50px; margin-top:5px;" src=" https://static.igem.org/mediawiki/2014/9/95/Freibur2014_pfeilrechts.png"> <!-- Pfeil fw--></a></div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 03:24, 18 October 2014
The Light System
Light inducible systems allow spatial and temporal control over gene expression. Our goal is the expression of the mCAT-1 receptor through a light inducible system to allow spatiotemporal control of the virus entry into the target cells. We used two different systems: expression is induced by light of the blue and red wavelength range. To test the performance of the light systems, we used an assay where the cells express secreted embryonic alkaline phosphatase (SEAP). The amount of produced SEAP can be measured by its enzymatic activity. It breaks down the substrate para-nitrophenylphosphate to the yellow para-nitrophenol, which can be measured with a spectrophotometer.
Function of the Light System
Because the proper function of the light system is important for our project, we tested two different light systems. The red light system is based on the protein phytochrome B (PhyB) from Arabidopsis thaliana and is activated by light around 660 nm, whereas the blue light system contains a protein from Avena sativa (AsLOV2) which requires 465 nm light for activation.
To test the function of the red light system, we used Chinese hamster ovary cells (CHO) that are known to work better for this purpose than other cell lines. For the blue light system, we tested CHO cells and human embryonic kidney cells (HEK-293T). As reporter the gene seap was transfected together with the genes required for the implementation of the blue and red light inducible systems. A SEAP-assay was performed after illumination. Measuring SEAP concentrations in the supernatant lead us two two important conclusions: First, the blue light system works more efficient than the red light system; second, the blue light system works better with HEK-293T cells than with CHO cells (Fig. 1).
The plasmids required for the implementation of the light inducible SEAP expression are:
Red light system:- pKM022: SV40_PhyB-VP16_IRES_TetR-PIF16
constitutive expression of PhyB-VP16 and TetR-PIF6 - pKM006: tetO-CMVmin_SEAP
SEAP under control of the tet operon
- pKM292: SV40_Gal4BD-LOV
constitutive expression of Gal4BD-LOV - pKM297: SV40_ePDZ-VP16
constitutive expression of ePDZ-VP16 - pKM084: Gal4UAS-CMVmin_SEAP
SEAP under control of the Gal4 upstream activating sequence

Figure 1: Comparison of the blue light system in CHO cells and in HEK-293T cells and comparison to the red light system in CHO cells.
To test the efficiency of the light system, SEAP expression after illumination was determined. The SEAP assay was performed 24 hours after light exposure. Labjournal

Figure 2: Efficiency of the blue light system using different time intervals of illumination.
To test the efficiency of the light system, SEAP expression after illumination was determined. The SEAP assay was performed 24 hours after light exposure. Cells were incubated with blue light for 1 hour, 2.5 hours and 5 hours. Labjournal
The blue light system is effective in activating the reporter SEAP which was introduced into the cells before illumination by transfections. As we know that the duration of illuminating the system with blue light (452 nm) is critical for its efficiency, we tested various time intervals for illumination. Our results indicate that five hours of illumination lead to the highest level of SEAP expression (Fig. 2).
Another important finding was the specificity of the blue light system. In the dark controls, we found almost no activation of the SEAP reporter, leading to a very low background level in our system.
SEAP as a Reporter
In order to test the capability of the blue light system for pattern generation, we transfected HEK-293T cells with the appropriate plasmids (PKM292 and PKM297) using seap as a reporter gene (PKM084). Cells were transfected in suspension and seeded on 96 well plates afterwards. Single wells of these plates were covered with a photo mask that prevents exposure to light. All other wells on the plate were illuminated to induce SEAP expression. SEAP that was secreted into the culture medium leads to a colour change from red to yellow after addition of pNPP (chromogenic substrate) by its phosphorylation.
First we generated a coarse pattern realizing that scattered light also reached wells that were close to the illuminated ones, leading to an unclear pattern. However, the rough shape of the desired pattern is discernible (Fig. 3A, B).
Since it was essential to completely prevent light exposure on covered plates, we tested 96 well plates with optically separated wells. We could generate patterns with sharp contours (Fig. 3C, D).
The colour change of pNPP in cell culture medium could be measured at 405 nm in a plate reader. Setting a distinct threshold made pattern recognition easier.
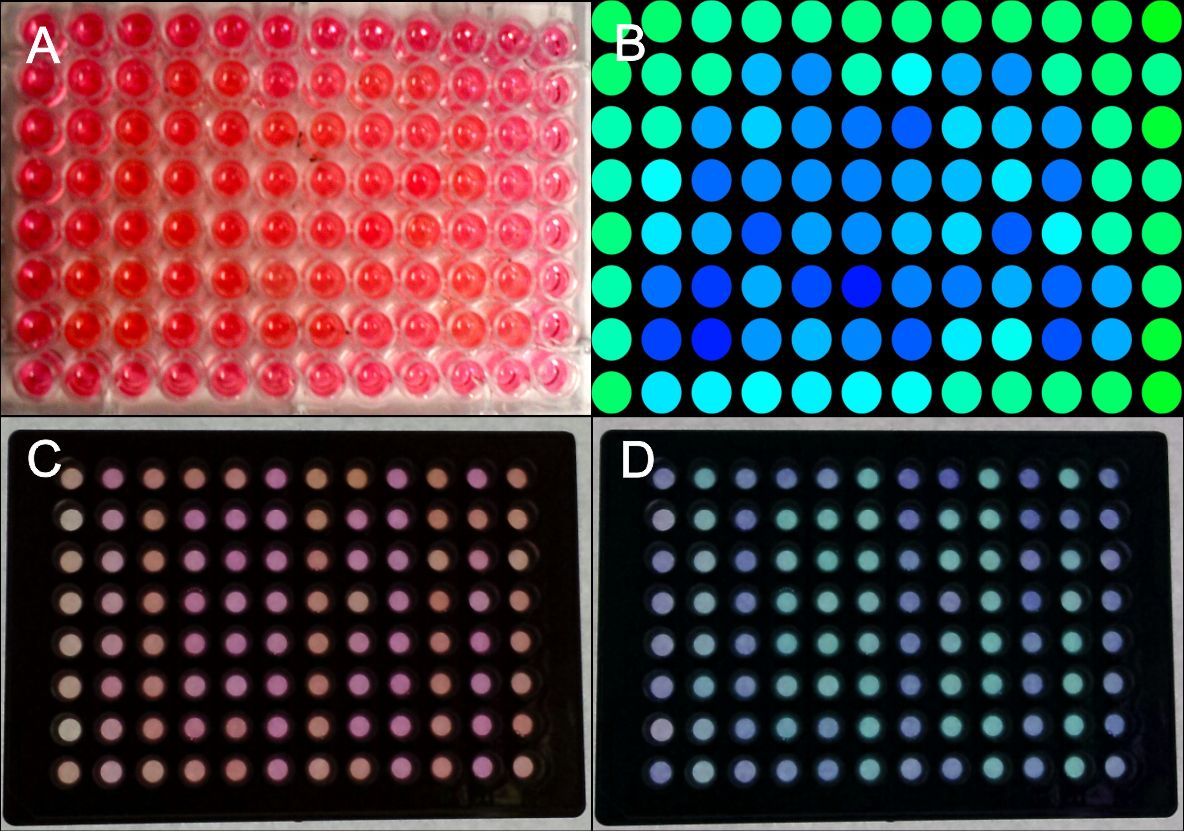
Figure 3: Generation of patterns on 96-well plates.
HEK-293T cells were transfected with the blue light system (PKM292, PKM297) and light induced SEAP as reporter (PKM084). Single wells were covered with a photo mask to prevent cell from light exposure. 24 hours after illumination with blue light, a SEAP assay was performed, leading to a colour change of wells containing SEAP. (A) Photo mask in heart form, (B) pseudocoloring done at a computer, (C) the same experiment was performed with optically separated wells, (D) the contrast was increased and the colours of the wells were changed at a computer. All wells together are forming the word: IGEM. Labjournal 1 Labjournal 2
For generating QR codes with our method 384-well plates were used. Since we had a collaboration with the iGEM Team Aachen 2014, they made a photo mask for a QR code for us fitting on these plates (Fig. 4). Collaborations.

Figure 4: Generation of a QR code on a 384 well plate.
HEK-293T cells were transfected with the blue light system (PKM292, PKM297) and light induced SEAP as reporter (PKM084). Single wells were covered with a photo mask to prevent cell from light exposure using a photo mask for a QR code (made by iGEM Team Aachen 2014). 24 hours after illumination with blue light a SEAP assay was performed leading to colour changing of wells containing SEAP. Only a part of the plate was used. (A) Photo mask for QR code generation, (B) Picture of the 384-well plate after the SEAP assay, (C) the same plate with contrast of the colours increased at a computer. Labjournal
 "
"

