Team:Oxford/Results
From 2014.igem.org
(Difference between revisions)
| Line 157: | Line 157: | ||
<br><br> | <br><br> | ||
<br> | <br> | ||
| - | + | <br> | |
<h1>Proteins can be targeted to microcompartments</h1> | <h1>Proteins can be targeted to microcompartments</h1> | ||
| Line 179: | Line 179: | ||
1. Microcompartment Shape Model: (see <a href="https://2014.igem.org/Team:Oxford/what_are_microcompartments?#show2" target="_blank">'Predicting the microcompartment structure'</a>) a model that simulates the effect of random point deviations in the microcompartment structure from the perfect icosahedral structure seen in carboxysomes.<br><br> | 1. Microcompartment Shape Model: (see <a href="https://2014.igem.org/Team:Oxford/what_are_microcompartments?#show2" target="_blank">'Predicting the microcompartment structure'</a>) a model that simulates the effect of random point deviations in the microcompartment structure from the perfect icosahedral structure seen in carboxysomes.<br><br> | ||
<img src=" https://static.igem.org/mediawiki/2014/7/7f/Microcompartment_shape_2.jpg " max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | <img src=" https://static.igem.org/mediawiki/2014/7/7f/Microcompartment_shape_2.jpg " max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | ||
| - | <strong>Figure | + | <strong>Figure 4</strong> |
<br> | <br> | ||
The output of the microcompartment shape model is a simulated microcompartment structure (blue) plotted alongside the perfect icosahedral structure of a carboxysome (red) which we used as a starting point for predicting the geometric structure of a microcompartment. | The output of the microcompartment shape model is a simulated microcompartment structure (blue) plotted alongside the perfect icosahedral structure of a carboxysome (red) which we used as a starting point for predicting the geometric structure of a microcompartment. | ||
| Line 189: | Line 189: | ||
" max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | " max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | ||
| - | <strong>Figure | + | <strong>Figure 5</strong> |
<br> | <br> | ||
We have plotted the trajectories of two different molecules along random, stochastically driven discretized paths. What we illustrate here is that the rate of collision (marked in red) is substantially increased through the use of spatial constraints. | We have plotted the trajectories of two different molecules along random, stochastically driven discretized paths. What we illustrate here is that the rate of collision (marked in red) is substantially increased through the use of spatial constraints. | ||
| Line 201: | Line 201: | ||
" max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | " max-height="500" style="float:right;position:relative; width:80%; margin-right:10%;margin-bottom:2%;margin-left:10%;"/><br> | ||
| - | <strong>Figure | + | <strong>Figure 6</strong> |
<br> | <br> | ||
Plotted here is the logarithmic relationship between star peptide bond length and reaction rate. This was simulated for Melbourne iGEM and suggests that the rate of reaction will be inversely proportional to average peptide length to the power of four. | Plotted here is the logarithmic relationship between star peptide bond length and reaction rate. This was simulated for Melbourne iGEM and suggests that the rate of reaction will be inversely proportional to average peptide length to the power of four. | ||
Revision as of 02:34, 18 October 2014
 "
"


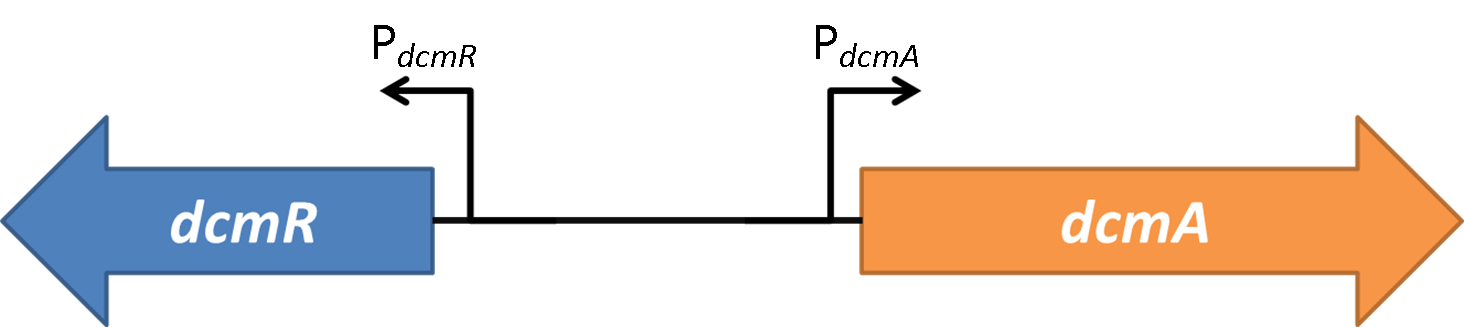





 Figure 2
Figure 2



























































