Team:Aachen/Project/Gal3
From 2014.igem.org
(→Natural Functions of Galectin-3) |
(→Natural Functions of Galectin-3) |
||
| Line 81: | Line 81: | ||
Galectins are proteins of the lectin family, which possess carbohydrate recognition domains binding specifically to β-galactoside sugar residues. In humans, 10 different galantines have been identified, among which is galectin-3. | Galectins are proteins of the lectin family, which possess carbohydrate recognition domains binding specifically to β-galactoside sugar residues. In humans, 10 different galantines have been identified, among which is galectin-3. | ||
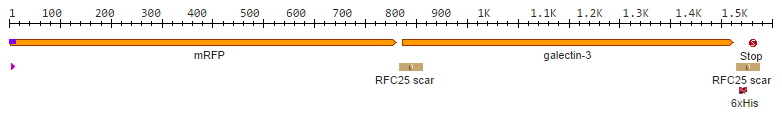
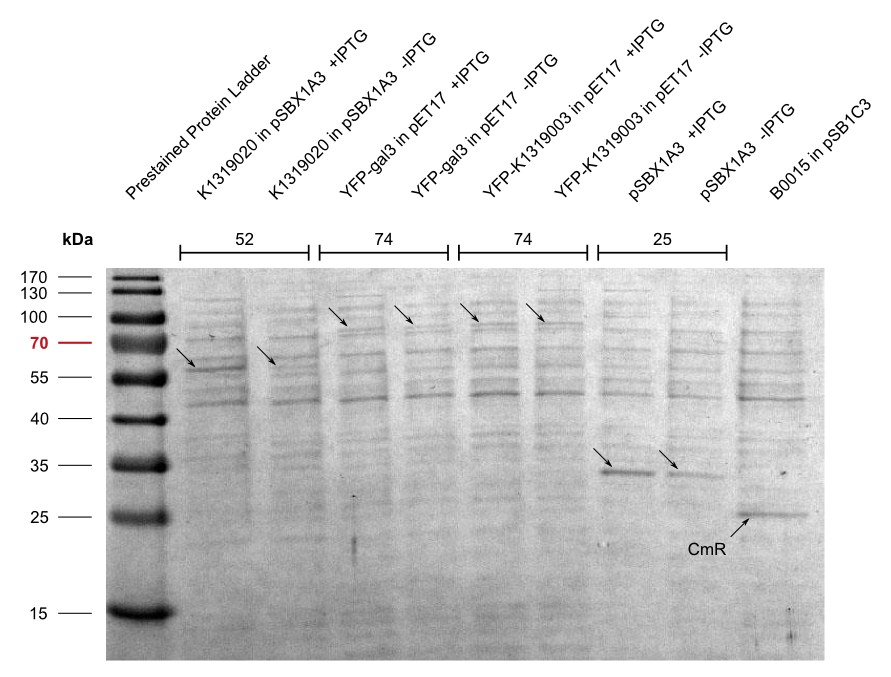
Galectin-3 has a size of about 31 kDA and is encoded by a single gene, LGALS3. It posesses many physiological functions, such as '''cell adhesion, cell growth and differentiation''', and contributes to the development of '''cancer, inflammation, fibrosis''' and others. | Galectin-3 has a size of about 31 kDA and is encoded by a single gene, LGALS3. It posesses many physiological functions, such as '''cell adhesion, cell growth and differentiation''', and contributes to the development of '''cancer, inflammation, fibrosis''' and others. | ||
| + | |||
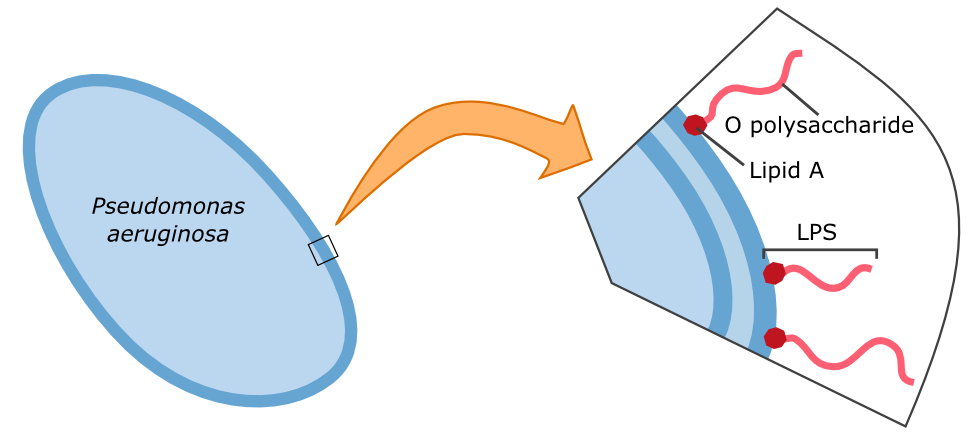
Human galectin-3 is a protein of the lectin-family that was shown to bind the LPS of multiple human pathogens. Some of them, including ''P. aeruginosa'' protect themselves against the human immune system by mimicking the lipopolysaccharides (LPS) present on human erythrocytes. | Human galectin-3 is a protein of the lectin-family that was shown to bind the LPS of multiple human pathogens. Some of them, including ''P. aeruginosa'' protect themselves against the human immune system by mimicking the lipopolysaccharides (LPS) present on human erythrocytes. | ||
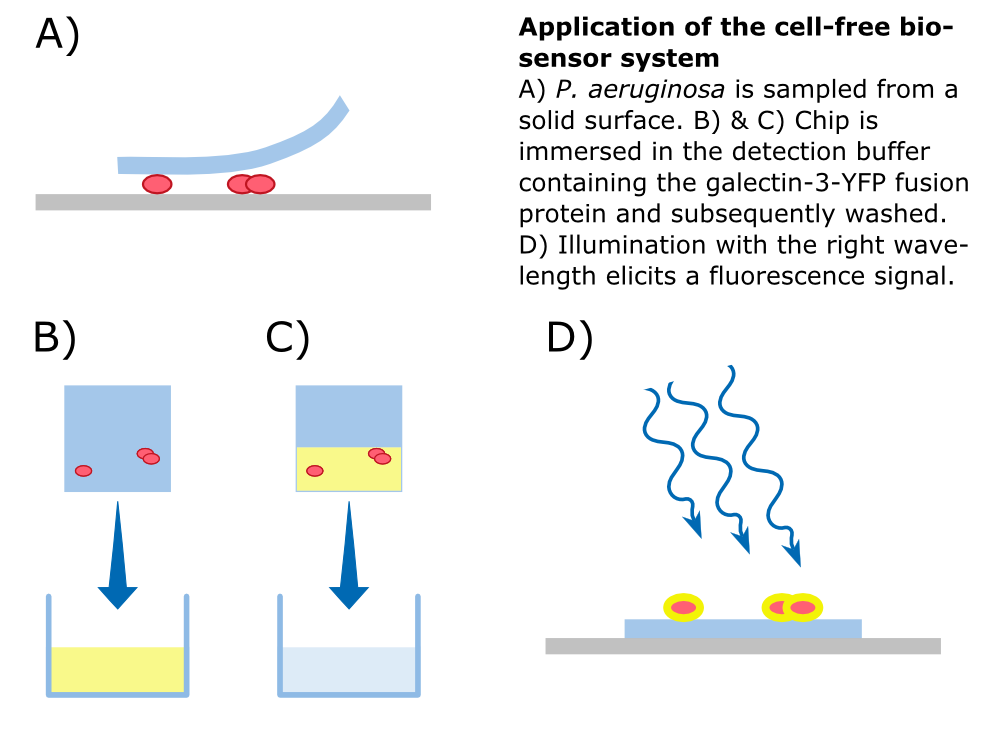
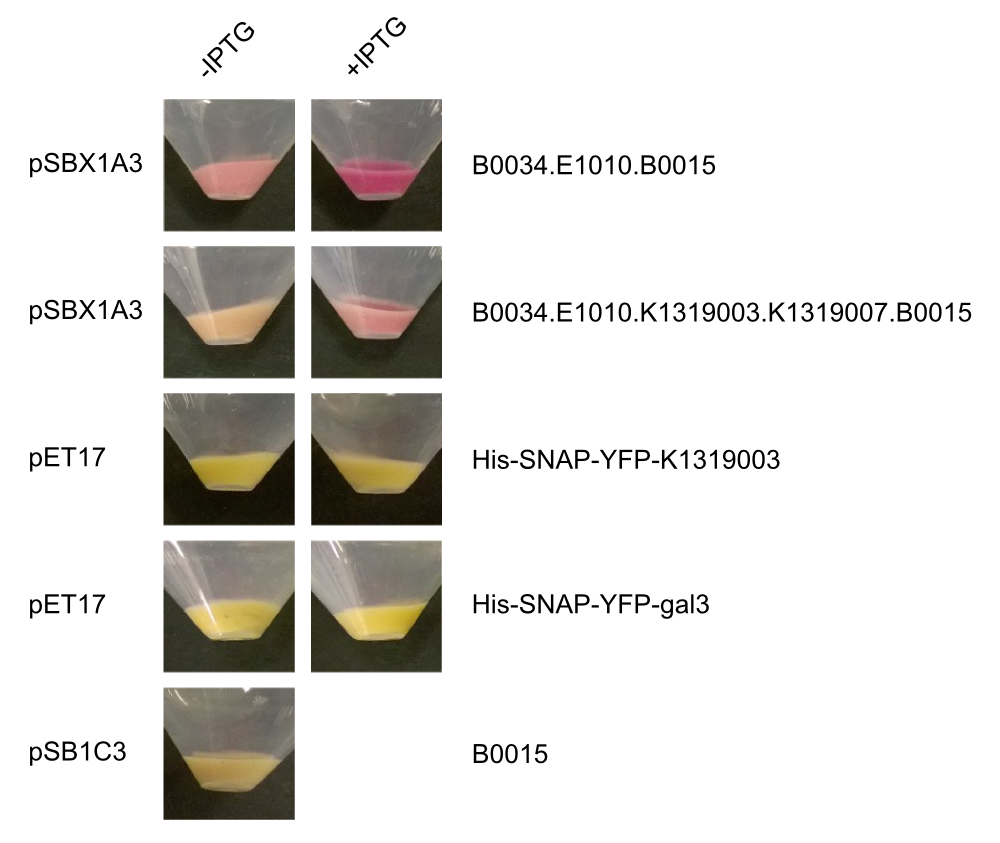
By making fusion proteins of galectin-3 with fluorescent reporter proteins, pathogens can be labelled and made visible by fluorescence-microscopy. | By making fusion proteins of galectin-3 with fluorescent reporter proteins, pathogens can be labelled and made visible by fluorescence-microscopy. | ||
Revision as of 01:19, 18 October 2014
|
|
|
|
 "
"