Team:Aachen/Notebook/Wetlab/July
From 2014.igem.org
(→31th) |
(→25th) |
||
| (24 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<ul class="menusmall-grid"> | <ul class="menusmall-grid"> | ||
| - | + | <!-- <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | |
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | ||
<div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | ||
| Line 14: | Line 14: | ||
</div> | </div> | ||
</a> | </a> | ||
| - | </li> | + | </li> --> |
<li style="width:106px;margin-left: 12px;margin-right: 12px;" > | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
| Line 91: | Line 91: | ||
* made chips with K1319042 and K731520 in NA and M9 medium. Images were taken every 30 min with the GelDoc | * made chips with K1319042 and K731520 in NA and M9 medium. Images were taken every 30 min with the GelDoc | ||
<center> | <center> | ||
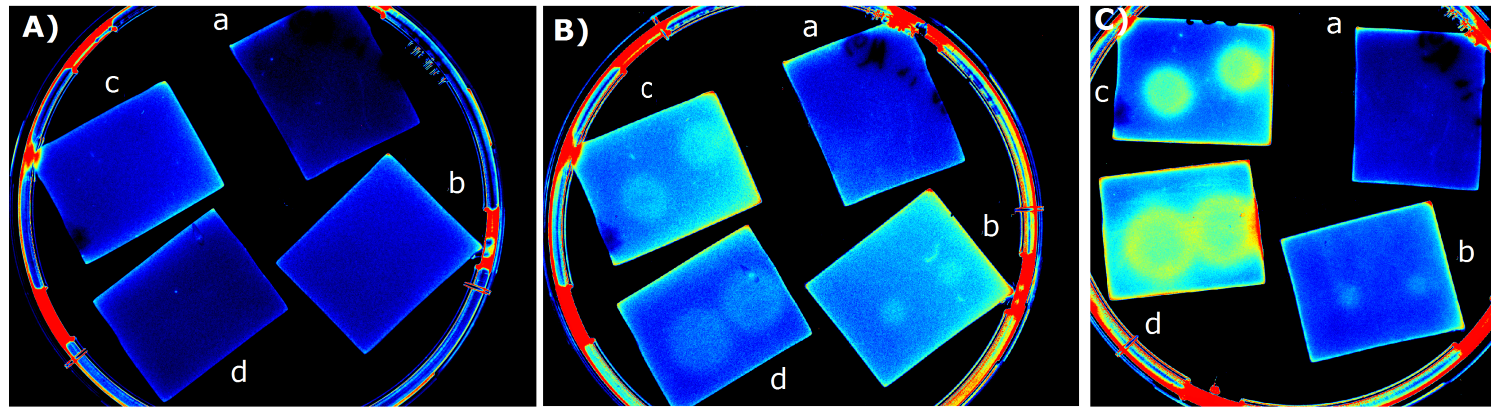
| - | {{Team:Aachen/Figure|Aachen_14-07-03_M9_iLOV_serie.png|title=Sensor Chips with K1319042 in M9 first try|subtitle=Sensor chips with K1319042 in M9 medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_14-07-03_M9_iLOV_serie.png|title=Sensor Chips with K1319042 in M9 first try|subtitle=Sensor chips with K1319042 in M9 medium with 1,5% agar. A) befor induction with IPTG B) 1 h after induction C) 4.5 h after induction a) induced with 2x 2 µl 1 mM IPTG b) induced with 2x 2 µl 10 mM IPTG c) induced with 2x 2 µl 0.1 mM IPTG d) induced with 2x 2 µl 100 mM IPTG|width=900px}} |
</center> | </center> | ||
* print K1319042 Chip on LB plate | * print K1319042 Chip on LB plate | ||
| Line 111: | Line 111: | ||
* made chips with K1319042 in M9, M9 + casamino acids, Hartman medium (HM). Images were taken every 30 min with the Geldoc | * made chips with K1319042 in M9, M9 + casamino acids, Hartman medium (HM). Images were taken every 30 min with the Geldoc | ||
<center> | <center> | ||
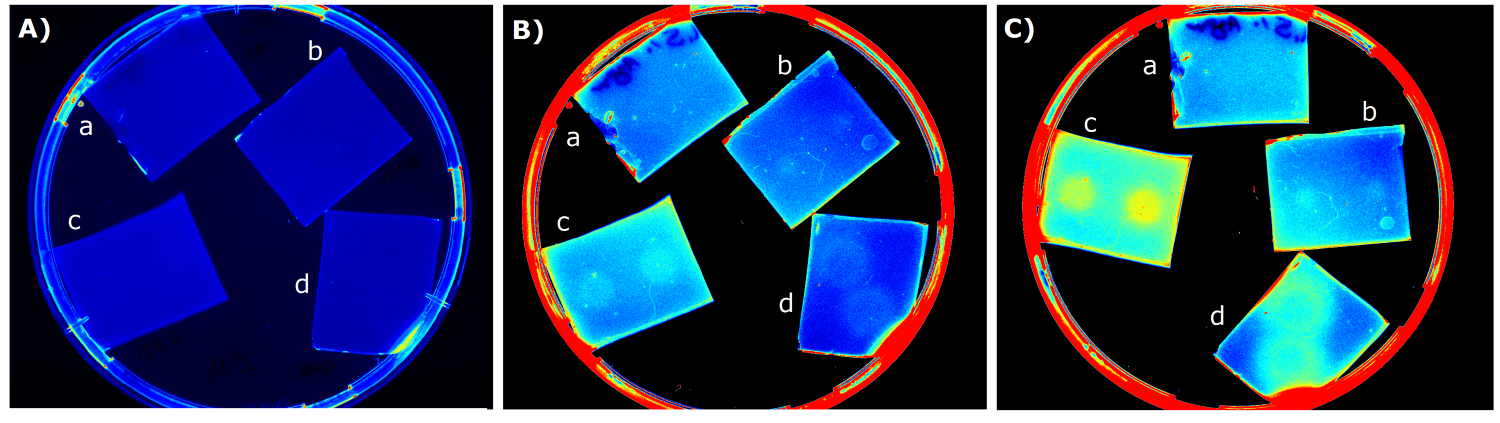
| - | {{Team:Aachen/Figure|Aachen_14-07-09_M9_iLOV_serie.png|title=Sensor Chips with K1319042 in M9 second try|subtitle=Sensor chips with K1319042 in M9 medium with 1,5 | + | {{Team:Aachen/Figure|Aachen_14-07-09_M9_iLOV_serie.png|title=Sensor Chips with K1319042 in M9 second try|subtitle=Sensor chips with K1319042 in M9 medium with 1,5% agar. A) befor induction with IPTG B) 1.5 h after induction C) 3 h after induction a) induced with 2x 2 µl 1 mM IPTG b) induced with 2x 2 µl 10 mM IPTG c) induced with 2x 2 µl 0.1 mM IPTG d) induced with 2x 2 µl 100 mM IPTG|width=900px}} |
</center> | </center> | ||
* made OmpA-linker_iLOV 3A-Assambley | * made OmpA-linker_iLOV 3A-Assambley | ||
| Line 139: | Line 139: | ||
***pSEVE641_BsFbFP pSEVA234_LasR | ***pSEVE641_BsFbFP pSEVA234_LasR | ||
***pSEVE641_BsFbFP pSB1C3_C0179 | ***pSEVE641_BsFbFP pSB1C3_C0179 | ||
| - | * made chips with K1319042 in Hartman, Hartman + 20 | + | * made chips with K1319042 in Hartman, Hartman + 20% glycerol, M9 medium images were taken every 30 min with the Geldoc |
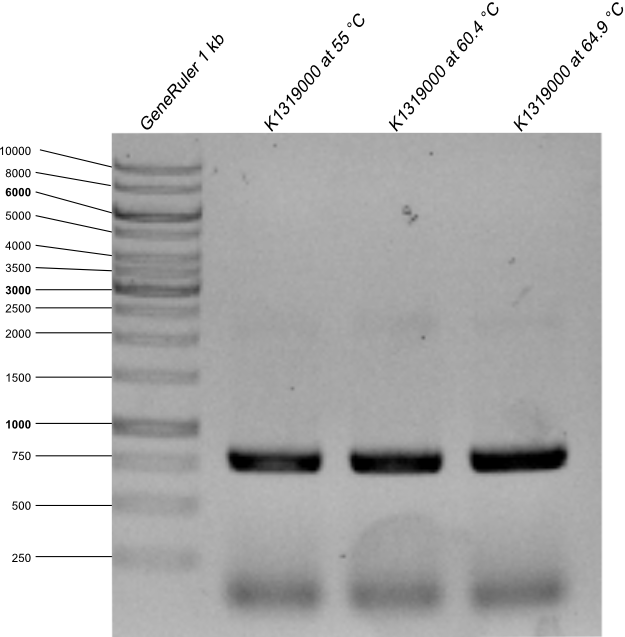
| + | * The PCR of Biobrick E0030 in vector pSB1C3 with primers for RFC 25 was made. The expected length is | ||
| + | ** forward primer EYFP_RFC25_F: ACGTTGAATTCGCGGCCGCTTCTAGATGGCCGGCGTGAGCAAGGGCGAGGAG | ||
| + | ** rewerse primer EYFP_RFC25_R: ATCCTGCAGCGGCCGCTACTAGTATTAACCGGTGTACAGCTCGTCCATGC | ||
| + | * PCR product gets number K1319000 | ||
| + | |||
| + | |||
| + | <center> | ||
| + | {| class="wikitable" | ||
| + | ! step !! temperature [°C] !! duration | ||
| + | |- | ||
| + | | denature || 94 || 60" | ||
| + | |- | ||
| + | | denature || 94 || 30" | ||
| + | |- | ||
| + | | anneal || 50/60,4/64,9 || 30" | ||
| + | |- | ||
| + | | elongate || 72 || 60" | ||
| + | |- | ||
| + | | elongate || 72 || 5' | ||
| + | |- | ||
| + | | store || 8 || indefinite | ||
| + | |} | ||
| + | </center> | ||
| + | |||
| + | * The agarose gel with PCR product was made. Expected length is 778 kb. | ||
| + | |||
| + | <center> | ||
| + | {{Team:Aachen/Figure|Aachen_14-09-17_E0030_with_RFC25.png|title=Agarose gel with PCR products after PCR from E0030 to K1319000 |width=400px}} | ||
| + | </center> | ||
| + | * as it can be seen on the picture PCR worked with all annealing temperatures, all samples were used for a transformation in DH5α after the restriction with DpnI. | ||
| + | |||
| + | == 17th == | ||
| + | * the PCR product was purificated and the concentration was measured. | ||
| + | * Restriktion of PCR product for the insert and E0030 for the vector with ''Eco''RI and PstI with following purification from agarose gel was made. | ||
| + | |||
| + | == 21st == | ||
| + | * the ligation of PCR product (K1319000) and the vector that was purified from agarose (from E0030) was made. | ||
| + | * ligated product was tranformed in DH5α. | ||
== 22nd == | == 22nd == | ||
| Line 147: | Line 185: | ||
== 23rd == | == 23rd == | ||
| - | * made 25 new HM+C plates with 1 | + | * made 25 new HM+C plates with 1% agar and 4 g/L glucose |
| - | * | + | * added 170 µL of the glucose stock (500 g/L) to the glucose-free HM+C-plates |
* 1 L of sterile HM+glucose was prepared | * 1 L of sterile HM+glucose was prepared | ||
* K1319042 was plated on HM+C+Glucose and HM+C+Glucose drops, 3x 5 mL HM+C+Glucose precultures were inoculated (17:00) | * K1319042 was plated on HM+C+Glucose and HM+C+Glucose drops, 3x 5 mL HM+C+Glucose precultures were inoculated (17:00) | ||
| Line 171: | Line 209: | ||
</center> | </center> | ||
| - | The powders for 1 L of 1x HM were dissolved in 20 mL of a 10 | + | The powders for 1 L of 1x HM were dissolved in 20 mL of a 10% w/v Casamino acid stock solution and filter-sterilized (.22 µm PES). To make agar chips, we can add 1 mL to the hot agar mixture, together with the three 500 µL 100x stocks for the HM. |
| - | We made 250 mL of HM+C+Glucose+Supplements plates with 1.5 | + | We made 250 mL of HM+C+Glucose+Supplements plates with 1.5% agar. |
Then we plated the following combinations: | Then we plated the following combinations: | ||
| Line 266: | Line 304: | ||
| J23101.E0240 || NEB10β || ++ || - || ++ | | J23101.E0240 || NEB10β || ++ || - || ++ | ||
|- | |- | ||
| - | | K1319042 || | + | | K1319042 || DH5α || ++ || - || (+) |
|- | |- | ||
| K131026 || NEB10β || ++ || - || - | | K131026 || NEB10β || ++ || - || - | ||
|- | |- | ||
| - | | K131026 || | + | | K131026 || DH5α || ++ || - || + |
|} | |} | ||
</center> | </center> | ||
| Line 281: | Line 319: | ||
== 29th == | == 29th == | ||
* inoculate 2x 150 ml cultures HM+Glucose+C | * inoculate 2x 150 ml cultures HM+Glucose+C | ||
| - | * prepared 200 ml 1.5 | + | * prepared 200 ml 1.5% agar |
* pre-cool centrifuge | * pre-cool centrifuge | ||
* centrifuge 1x 50, 1x 100 and 1x 150 ml | * centrifuge 1x 50, 1x 100 and 1x 150 ml | ||
| Line 299: | Line 337: | ||
<ul class="menusmall-grid"> | <ul class="menusmall-grid"> | ||
| - | + | <!-- <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | |
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Notebook/Wetlab/March" style="color:black"> | ||
<div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | <div class="menusmall-item menusmall-info" style="height:100px; width: 100px;" ><div class="menukachel" style="top: 30%; font-size: 16px;">Go to March</div></div> | ||
| Line 305: | Line 343: | ||
</div> | </div> | ||
</a> | </a> | ||
| - | </li> | + | </li> --> |
<li style="width:106px;margin-left: 12px;margin-right: 12px;" > | <li style="width:106px;margin-left: 12px;margin-right: 12px;" > | ||
Latest revision as of 16:16, 17 October 2014
|
 "
"