Team:Cornell/project/wetlab
From 2014.igem.org
(Difference between revisions)
| Line 6: | Line 6: | ||
$('li.p_wet_over').addClass('active'); | $('li.p_wet_over').addClass('active'); | ||
}); | }); | ||
| - | + | </script> | |
<body> | <body> | ||
| - | <div class="container"> | + | <div class="container" id="top"> |
<div class="row"> | <div class="row"> | ||
<div class="col-md-9 col-xs-15"> | <div class="col-md-9 col-xs-15"> | ||
| - | <h1>Idea</h1> | + | <h1>Idea</h1> |
Mercury, nickel, and lead were targeted for sequestration by our strain of bacteria by utilizing the efficient binding properties of yeast metallothionein (CRS5) and specificity of the respective metal transport proteins, <i>merT</i> and <i>merP</i><sup>1</sup>, <i>nixA</i><sup>2</sup>, and <i>CBP4</i>. The yeast metallothionein<sup>3</sup> and two heavy metal transport proteins were BioBricked. Our wetlab sub team co-transformed the parts using different plasmids and multiple selection markers to develop functional composite constructs. The idea of utilizing metallothioneins in parallel with metal transporters for sequestration has been studied in depth for mercury and nickel. However, this is a novel idea for lead. | Mercury, nickel, and lead were targeted for sequestration by our strain of bacteria by utilizing the efficient binding properties of yeast metallothionein (CRS5) and specificity of the respective metal transport proteins, <i>merT</i> and <i>merP</i><sup>1</sup>, <i>nixA</i><sup>2</sup>, and <i>CBP4</i>. The yeast metallothionein<sup>3</sup> and two heavy metal transport proteins were BioBricked. Our wetlab sub team co-transformed the parts using different plasmids and multiple selection markers to develop functional composite constructs. The idea of utilizing metallothioneins in parallel with metal transporters for sequestration has been studied in depth for mercury and nickel. However, this is a novel idea for lead. | ||
<br> <br> | <br> <br> | ||
To test the sequestration efficiency of each metal, <i>E.coli</i> BL21-A1 was transformed with the yeast metallothionein as well the respective metal transporter for the targeted metal. The sequestering strains can be placed into fiber reactors to develop functional sequestering filters as part of the dry lab portion of our project. | To test the sequestration efficiency of each metal, <i>E.coli</i> BL21-A1 was transformed with the yeast metallothionein as well the respective metal transporter for the targeted metal. The sequestering strains can be placed into fiber reactors to develop functional sequestering filters as part of the dry lab portion of our project. | ||
| - | |||
</div> | </div> | ||
| - | <div class="col-md-3 col-xs-3"> | + | <div class="col-md-3 col-xs-3"> |
| - | < | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/9/91/Protein.PNG"> |
| - | </div> | + | <img src="https://static.igem.org/mediawiki/2014/9/91/Protein.PNG"> |
| + | </a> | ||
| + | </div> | ||
</div> | </div> | ||
| - | |||
<div class="row"> | <div class="row"> | ||
<div class="col-md-12 col-xs-18"> | <div class="col-md-12 col-xs-18"> | ||
<h1>Components</h1> | <h1>Components</h1> | ||
| - | <center><img src="https://static.igem.org/mediawiki/2014/thumb/1/16/Componenttable.PNG/800px-Componenttable.PNG | + | <center><img src="https://static.igem.org/mediawiki/2014/thumb/1/16/Componenttable.PNG/800px-Componenttable.PNG"></center> |
| - | + | ||
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
<div class="col-md-12 col-xs-18"> | <div class="col-md-12 col-xs-18"> | ||
<h1>Experiments</h1> | <h1>Experiments</h1> | ||
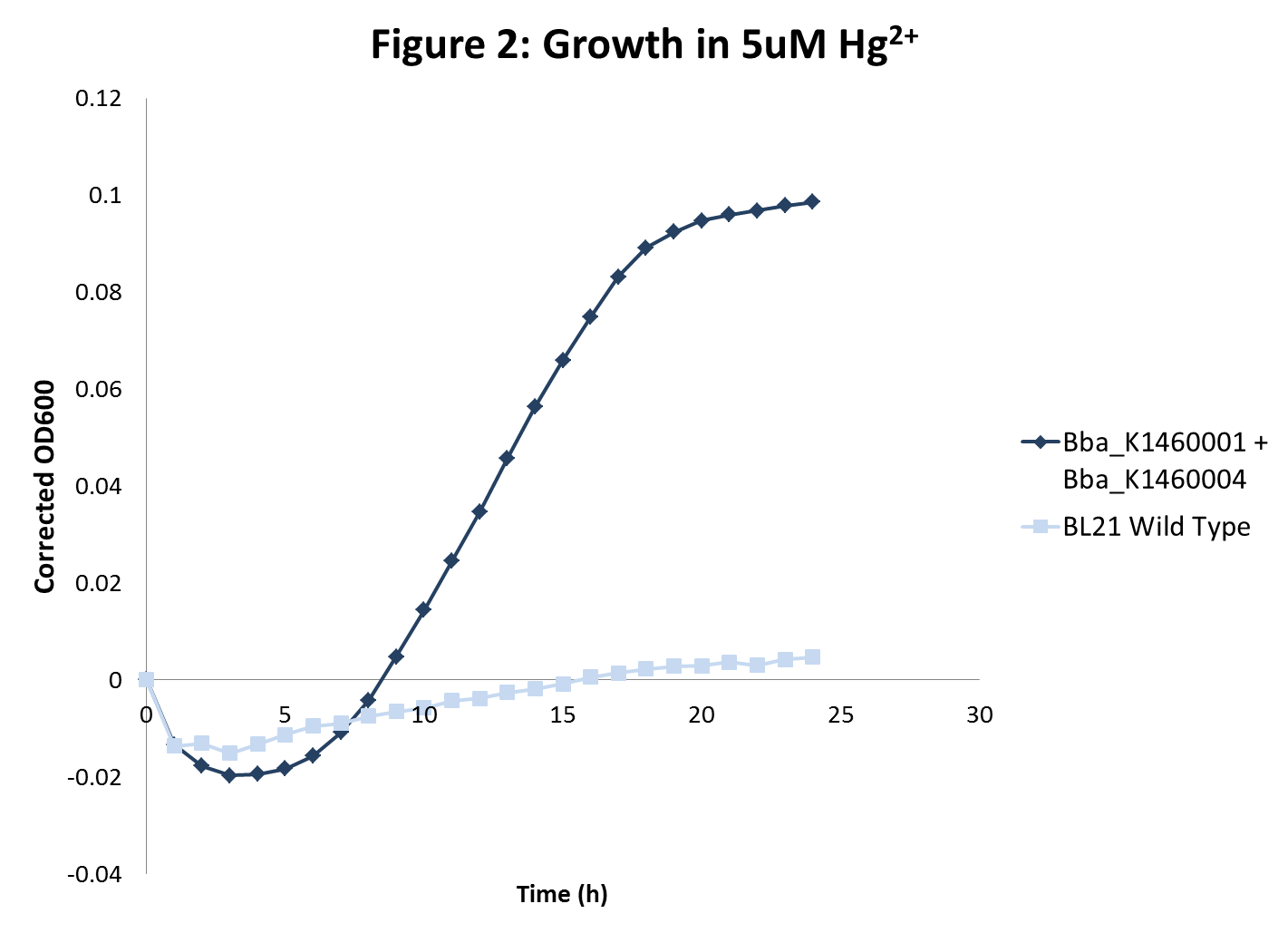
The growth rates of the sequestering strain were measured using spectrophotometry. In addition, two methods were used to determine heavy metal sequestration efficiency. <br><br> | The growth rates of the sequestering strain were measured using spectrophotometry. In addition, two methods were used to determine heavy metal sequestration efficiency. <br><br> | ||
| - | + | <ol> | |
| - | <ol> | + | <li> Spectrophotometer was used to analyze and compare the kinetic growth rates of E. coli cultures expressing only the metallothionein protein, only the transporter proteins, both metallothionein and transporter proteins, and just the vector backbone as a control. </li> |
| - | <li> Spectrophotometer was used to analyze and compare the kinetic growth rates of E. coli cultures expressing only the metallothionein protein, only the transporter proteins, both metallothionein and transporter proteins, and just the vector backbone as a control. </li> | + | <ul> |
| - | <ul> | + | <li>We hypothesized that the control culture would be mildly sensitive to growth in metal-containing media, while the cultures with the transport protein would be more sensitive to growth in metal-containing media due to increased access of the heavy metal to cellular machinery. Finally, bacteria transformed with both the metal transporter and metallothionein protein would be the least sensitive in metal-containing media. Each strain was grown in different heavy metal concentrations.</li> |
| - | <li>We hypothesized that the control culture would be mildly sensitive to growth in metal-containing media, while the cultures with the transport protein would be more sensitive to growth in metal-containing media due to increased access of the heavy metal to cellular machinery. Finally, bacteria transformed with both the metal transporter and metallothionein protein would be the least sensitive in metal-containing media. Each strain was grown in different heavy metal concentrations.</li> | + | </ul> |
| - | </ul> | + | <br> |
| - | <li>Sequestration efficiency was measured by growing both the wild type strains as well as sequestering strains in different concentrations of heavy metals. The concentration of heavy metals after growth was measured in two ways: </li> | + | <li>Sequestration efficiency was measured by growing both the wild type strains as well as sequestering strains in different concentrations of heavy metals. The concentration of heavy metals after growth was measured in two ways: </li> |
| - | <ul> | + | <ul> |
| - | <li> Nutrient Analysis Lab at Cornell University</li> | + | <li> Nutrient Analysis Lab at Cornell University</li> |
| - | <li> Using green-fluorescent heavy metal indicator Phen Green</li> | + | <li> Using green-fluorescent heavy metal indicator Phen Green</li> |
| - | </ul> | + | </ul> |
| - | </ol | + | </ol> |
| - | + | ||
</div> | </div> | ||
</div> | </div> | ||
| - | + | <div class = "row"> | |
| - | + | <div class="col-md-12 col-xs-18"> | |
| - | <div class = "row"> | + | <h1>Results</h1> |
| - | <div class="col-md-12 col-xs-18"> | + | <ol> |
| - | <h1>Results</h1> | + | <li>BL21 strains with the merT/merP transporters showed increased sensitivity to mercury concentrations as expected. In addition, in high mercury concentrations, wild type strain growth was inhibited more than sequestration strain growths were inhibited. However, there was no inhibition of growth with wild type BL21 or strains with transporters for lead and nickel even at very high metal concentrations. </li> |
| - | <ol> | + | </ol> |
| - | <li>BL21 strains with the merT/merP transporters showed increased sensitivity to mercury concentrations as expected. In addition, in high mercury concentrations, wild type strain growth was inhibited more than sequestration strain growths were inhibited. However, there was no inhibition of growth with wild type BL21 or strains with transporters for lead and nickel even at very high metal concentrations. </li> | + | </div> |
| - | </ol> | + | |
| - | + | ||
| - | <div | + | |
<div class="col-md-6 col-xs-9"> | <div class="col-md-6 col-xs-9"> | ||
| - | < | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/8/82/Cornell_merTandmerP_growth.png"> |
| - | </ | + | <img src="https://static.igem.org/mediawiki/2014/8/82/Cornell_merTandmerP_growth.png"> |
| - | + | </a> | |
| - | <div class="col-md-6 col-xs-9"> | + | </div> |
| - | < | + | <div class="col-md-6 col-xs-9"> |
| - | </ | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/1/1c/Cornell_merandmet_growth.png"> |
| - | </div> | + | <img src="https://static.igem.org/mediawiki/2014/1/1c/Cornell_merandmet_growth.png"> |
| - | + | </a> | |
| - | < | + | </div> |
| - | + | </div> | |
| - | <div class = "row"> | + | <div class = "row"> |
| - | <div class="col-md-12 col-xs-18"> | + | <div class="col-md-12 col-xs-18"> |
| - | <ol start = "2"> | + | <ol start = "2"> |
| - | <li>Regarding cell density, both lead and nickel sequestering strains removed significantly more metal compared to the wild type BL21 strain. The mercury sequestering strain did not sequester significantly more metal compared to the wild type strain.</li> | + | <li>Regarding cell density, both lead and nickel sequestering strains removed significantly more metal compared to the wild type BL21 strain. The mercury sequestering strain did not sequester significantly more metal compared to the wild type strain.</li> |
| - | </ol> | + | </ol> |
| - | + | </div> | |
| - | + | </div> | |
| - | </div> | + | <div class="row"> |
| - | </div> | + | |
| - | <div class="row"> | + | |
<div class="col-md-4 col-xs-6"> | <div class="col-md-4 col-xs-6"> | ||
| - | < | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/5/57/Cornell_Lead_per_OD.png"> |
| - | + | <img src="https://static.igem.org/mediawiki/2014/5/57/Cornell_Lead_per_OD.png"> | |
| - | < | + | </a> |
| - | + | <a href="https://2014.igem.org/Team:Cornell/project/wetlab/lead"> | |
| - | < | + | <button type="button" class="btn btn-default btn-lg center"> |
| - | + | Lead Data | |
| - | </ | + | </button> |
| - | </ | + | </a> |
</div> | </div> | ||
<div class="col-md-4 col-xs-6"> | <div class="col-md-4 col-xs-6"> | ||
| - | < | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/1/1c/Cornell_merandmet_growth.png"> |
| - | + | <img src="https://static.igem.org/mediawiki/2014/1/1d/Cornell_mercury_per_OD.png"> | |
| - | < | + | </a> |
| - | + | <a href="https://2014.igem.org/Team:Cornell/project/wetlab/mercury"> | |
| - | < | + | <button type="button" class="btn btn-default btn-lg center"> |
| - | + | Mercury Data | |
| - | </ | + | </button> |
| - | </ | + | </a> |
</div> | </div> | ||
<div class="col-md-4 col-xs-6"> | <div class="col-md-4 col-xs-6"> | ||
| - | + | <a class="thumbnail" data-toggle="lightbox" href="https://static.igem.org/mediawiki/2014/c/ca/Cornell_nickel_per_OD.png"> | |
| - | + | <img src="https://static.igem.org/mediawiki/2014/c/ca/Cornell_nickel_per_OD.png"> | |
| - | <img src="https://static.igem.org/mediawiki/2014/c/ca/Cornell_nickel_per_OD.png | + | </a> |
| - | + | <a href="https://2014.igem.org/Team:Cornell/project/wetlab/nickel"> | |
| - | + | <button type="button" class="btn btn-default btn-lg center"> | |
| - | < | + | Nickel Data |
| - | + | </button> | |
| - | </ | + | </a> |
| - | </ | + | |
</div> | </div> | ||
</div> | </div> | ||
| - | <div class="row"> | + | <div class="row"> |
<div class="col-md-12 col-xs-18"> | <div class="col-md-12 col-xs-18"> | ||
<h1 style="margin-bottom: 0px">References</h1> | <h1 style="margin-bottom: 0px">References</h1> | ||
| Line 120: | Line 109: | ||
<ol> | <ol> | ||
<li>Wilson, D. B. Construction and characterization of Escherichia coli genetically engineered for Construction and Characterization of Escherichia coli Genetically Engineered for Bioremediation of Hg 2 ϩ -Contaminated Environments. 2–6 (1997).</li> | <li>Wilson, D. B. Construction and characterization of Escherichia coli genetically engineered for Construction and Characterization of Escherichia coli Genetically Engineered for Bioremediation of Hg 2 ϩ -Contaminated Environments. 2–6 (1997).</li> | ||
| - | <li>Krishnaswamy, R. & Wilson, D. B. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl. Environ. Microbiol. 66, 5383–6 (2000).</li> | + | <li>Krishnaswamy, R. & Wilson, D. B. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl. Environ. Microbiol. 66, 5383–6 (2000).</li> |
| - | <li>Huang, J. et al. Fission yeast HMT1 lowers seed cadmium through phytochelatin-dependent vacuolar sequestration in Arabidopsis. Plant Physiol. 158, 1779–88 (2012).</li> | + | <li>Huang, J. et al. Fission yeast HMT1 lowers seed cadmium through phytochelatin-dependent vacuolar sequestration in Arabidopsis. Plant Physiol. 158, 1779–88 (2012).</li> |
| - | + | </ol> | |
</div> | </div> | ||
</div> | </div> | ||
Revision as of 06:58, 16 October 2014
Wet Lab
Idea
Mercury, nickel, and lead were targeted for sequestration by our strain of bacteria by utilizing the efficient binding properties of yeast metallothionein (CRS5) and specificity of the respective metal transport proteins, merT and merP1, nixA2, and CBP4. The yeast metallothionein3 and two heavy metal transport proteins were BioBricked. Our wetlab sub team co-transformed the parts using different plasmids and multiple selection markers to develop functional composite constructs. The idea of utilizing metallothioneins in parallel with metal transporters for sequestration has been studied in depth for mercury and nickel. However, this is a novel idea for lead.To test the sequestration efficiency of each metal, E.coli BL21-A1 was transformed with the yeast metallothionein as well the respective metal transporter for the targeted metal. The sequestering strains can be placed into fiber reactors to develop functional sequestering filters as part of the dry lab portion of our project.
Components

Experiments
The growth rates of the sequestering strain were measured using spectrophotometry. In addition, two methods were used to determine heavy metal sequestration efficiency.- Spectrophotometer was used to analyze and compare the kinetic growth rates of E. coli cultures expressing only the metallothionein protein, only the transporter proteins, both metallothionein and transporter proteins, and just the vector backbone as a control.
- We hypothesized that the control culture would be mildly sensitive to growth in metal-containing media, while the cultures with the transport protein would be more sensitive to growth in metal-containing media due to increased access of the heavy metal to cellular machinery. Finally, bacteria transformed with both the metal transporter and metallothionein protein would be the least sensitive in metal-containing media. Each strain was grown in different heavy metal concentrations.
- Sequestration efficiency was measured by growing both the wild type strains as well as sequestering strains in different concentrations of heavy metals. The concentration of heavy metals after growth was measured in two ways:
- Nutrient Analysis Lab at Cornell University
- Using green-fluorescent heavy metal indicator Phen Green
Results
- BL21 strains with the merT/merP transporters showed increased sensitivity to mercury concentrations as expected. In addition, in high mercury concentrations, wild type strain growth was inhibited more than sequestration strain growths were inhibited. However, there was no inhibition of growth with wild type BL21 or strains with transporters for lead and nickel even at very high metal concentrations.
- Regarding cell density, both lead and nickel sequestering strains removed significantly more metal compared to the wild type BL21 strain. The mercury sequestering strain did not sequester significantly more metal compared to the wild type strain.
References
- Wilson, D. B. Construction and characterization of Escherichia coli genetically engineered for Construction and Characterization of Escherichia coli Genetically Engineered for Bioremediation of Hg 2 ϩ -Contaminated Environments. 2–6 (1997).
- Krishnaswamy, R. & Wilson, D. B. Construction and characterization of an Escherichia coli strain genetically engineered for Ni(II) bioaccumulation. Appl. Environ. Microbiol. 66, 5383–6 (2000).
- Huang, J. et al. Fission yeast HMT1 lowers seed cadmium through phytochelatin-dependent vacuolar sequestration in Arabidopsis. Plant Physiol. 158, 1779–88 (2012).
 "
"