Team:IvyTech SouthBend IN/progress
From 2014.igem.org
(Difference between revisions)
| Line 164: | Line 164: | ||
<div id="memo3g"><a name="91420">Week</a> of 9/14/14 through 9/20/14</div><br /> | <div id="memo3g"><a name="91420">Week</a> of 9/14/14 through 9/20/14</div><br /> | ||
<ul> | <ul> | ||
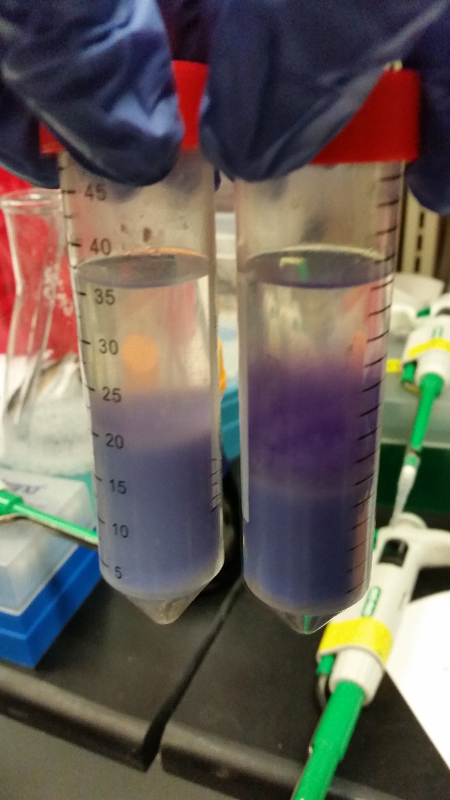
| - | <li>This week we began our attempts on the Beta-galactosidase activity assay. We plan to run this assay three different ways. We will first complete it using intact cells, measuring the activity that is produce inside the cell. Then we will use chemically lysed cells. By lysing the cells in this way, we are ensuring a higher accuracy in regards to amount of cells being lysed. The chemicals will destroy the cell wall and the beta-gal | + | <li>This week we began our attempts on the Beta-galactosidase activity assay. We plan to run this assay three different ways. We will first complete it using intact cells, measuring the activity that is produce inside the cell. Then we will use chemically lysed cells. By lysing the cells in this way, we are ensuring a higher accuracy in regards to amount of cells being lysed. The chemicals will destroy the cell wall and the beta-gal complementation can occur in vitro.</li><br /> |
<li>Finally, we plan to bring the core idea of our research project together, and then we will run this assay using cells that have been lysed with bacteriophage.</li><br /> | <li>Finally, we plan to bring the core idea of our research project together, and then we will run this assay using cells that have been lysed with bacteriophage.</li><br /> | ||
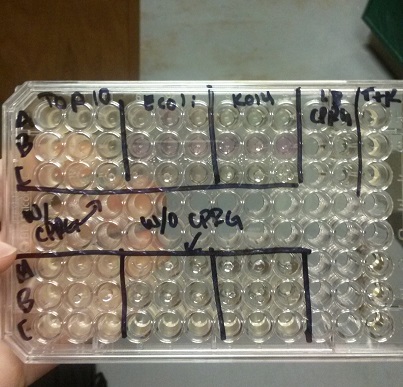
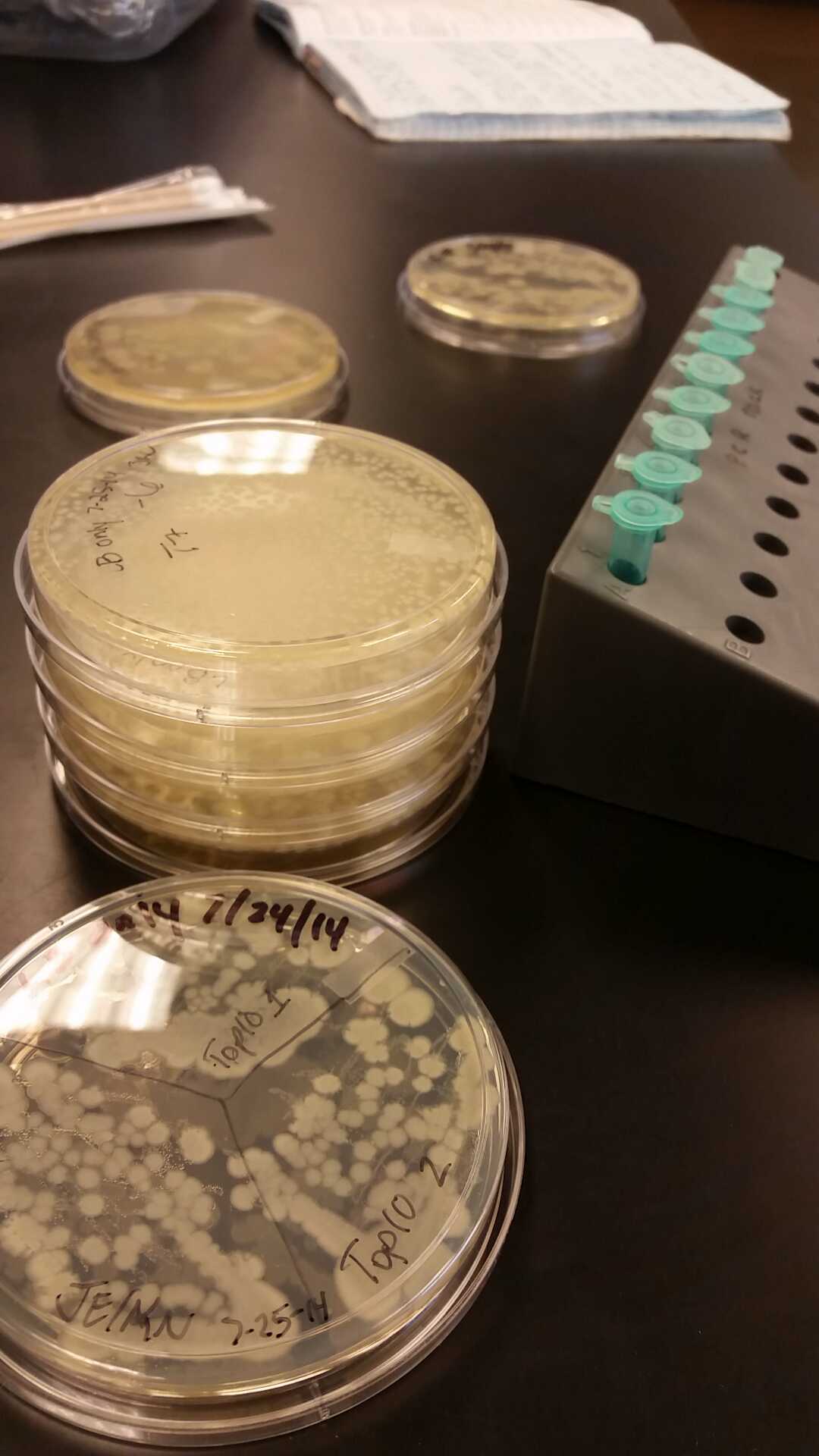
<li>In all of these, we will be using chlorophenol red (CPRG) as the substrate. This will cause a color change that ranges from yellow to dark red. We are using Top 10 E. coli cells as our negative control. These only produce one out of the two parts that make up the enzyme. We will be using E. coli C as our positive control. We are testing the activity produced in our part K1477014 transformed into Top 10 cells.</li><br /> | <li>In all of these, we will be using chlorophenol red (CPRG) as the substrate. This will cause a color change that ranges from yellow to dark red. We are using Top 10 E. coli cells as our negative control. These only produce one out of the two parts that make up the enzyme. We will be using E. coli C as our positive control. We are testing the activity produced in our part K1477014 transformed into Top 10 cells.</li><br /> | ||
| Line 340: | Line 340: | ||
<div id="memo3g"><a name="72026">Week</a> of 7/20/14 through 7/26/14</div><br /> | <div id="memo3g"><a name="72026">Week</a> of 7/20/14 through 7/26/14</div><br /> | ||
<ul> | <ul> | ||
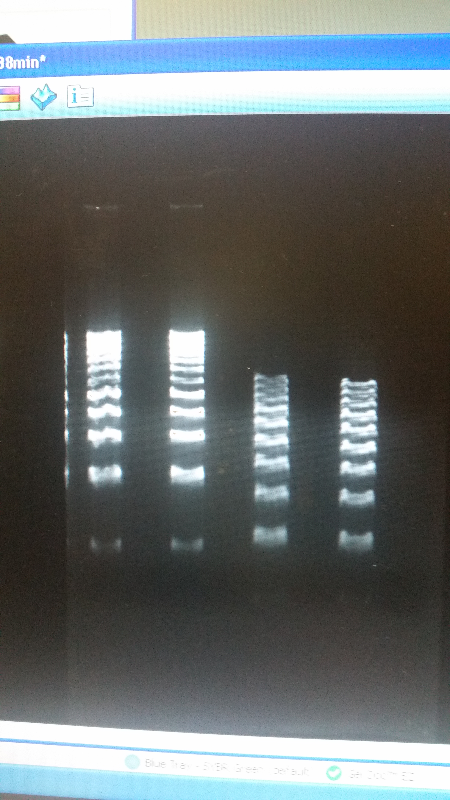
| - | <li>We have created a stock of our two parts on the pSB1C3 backbone. We purified both parts, will prepare to send them in to iGEM by running an electrophoresis gel to prove that our parts are what we say they are. We ran two gels, but the DNA bands are coming out deformed for some reason and the DNA ladder does not have distinguished bands that can be used to count how many | + | <li>We have created a stock of our two parts on the pSB1C3 backbone. We purified both parts, will prepare to send them in to iGEM by running an electrophoresis gel to prove that our parts are what we say they are. We ran two gels, but the DNA bands are coming out deformed for some reason and the DNA ladder does not have distinguished bands that can be used to count how many kilo-base pairs our part has. We are looking into why this is. Possible explanations include incorrect voltage, incorrect agarose gel thickness, or incorrect amount of DNA added. Adjustments will be made in the future.</li><br /> |
<li>We have also performed 3A biobrick assembly two more times and have a new stock of our part on its original backbone pSB1AT3.</li><br /> | <li>We have also performed 3A biobrick assembly two more times and have a new stock of our part on its original backbone pSB1AT3.</li><br /> | ||
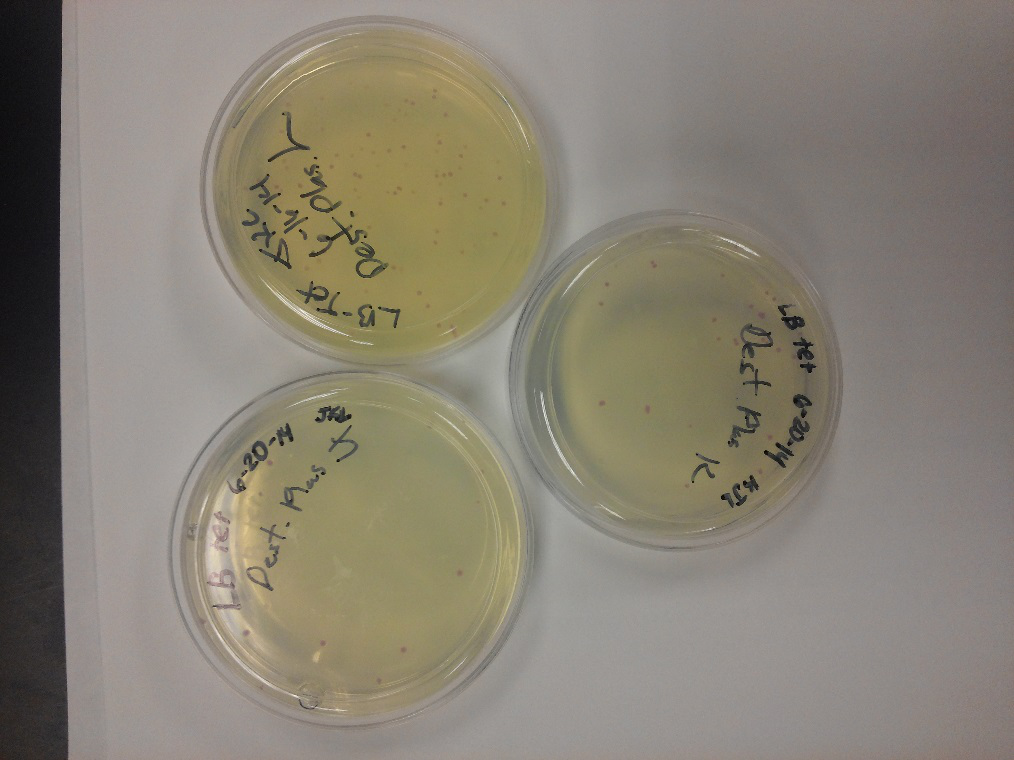
<li>Our problem of contamination is continuing to persist. During our attempts of recreating a new stock of our parts, we found bright yellow colonies all over more than half of the plates we streaked.</li> | <li>Our problem of contamination is continuing to persist. During our attempts of recreating a new stock of our parts, we found bright yellow colonies all over more than half of the plates we streaked.</li> | ||
| Line 363: | Line 363: | ||
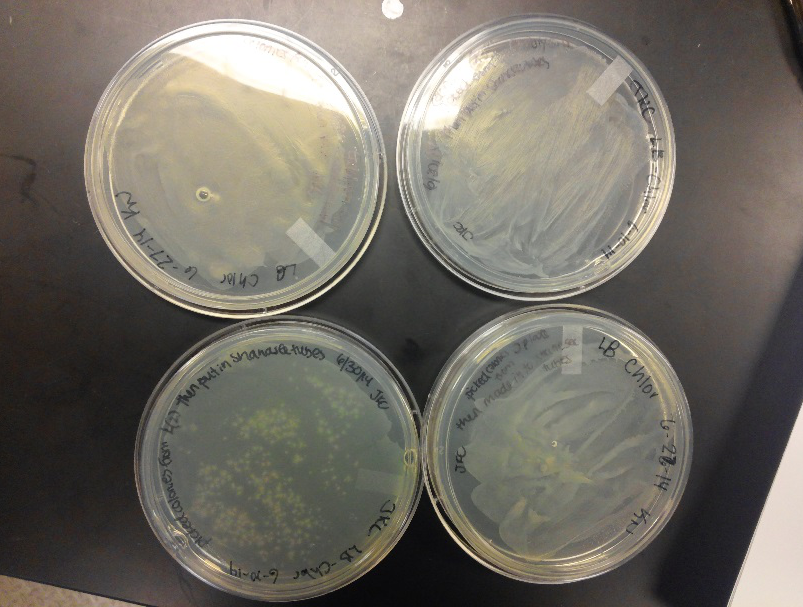
<li>This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.</li><br /> | <li>This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.</li><br /> | ||
<li>Unfortunately, We no longer have any stock of our original part. So, now we are again following the 3A Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.</li> <br /> | <li>Unfortunately, We no longer have any stock of our original part. So, now we are again following the 3A Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.</li> <br /> | ||
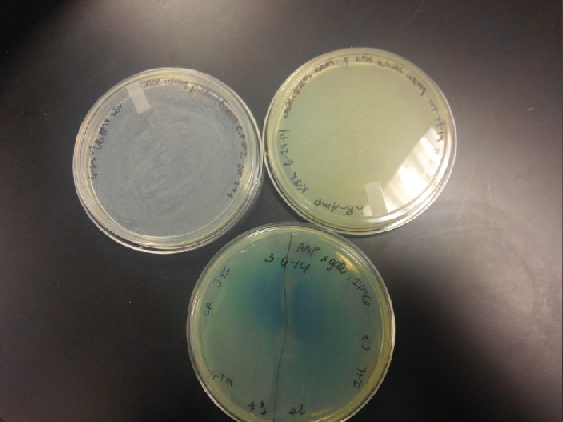
| - | <li>Our second run through of the protocol resulted in the growth of red colonies on chlor plates after | + | <li>Our second run through of the protocol resulted in the growth of red colonies on chlor plates after electroporation of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.</li> |
</ul> | </ul> | ||
<!-- column 1 end --> | <!-- column 1 end --> | ||
| Line 402: | Line 402: | ||
<ul> | <ul> | ||
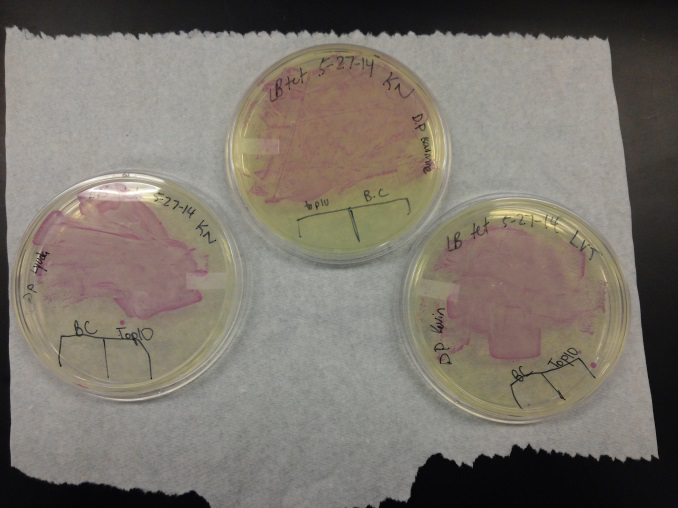
<li>Last week Lyuda and Kevin ligated the part K1477014 with J23102(constitutive promoter) I732020( LacZoperon Mutant) into pSB1AT3(destination plasmid).</li><br /> | <li>Last week Lyuda and Kevin ligated the part K1477014 with J23102(constitutive promoter) I732020( LacZoperon Mutant) into pSB1AT3(destination plasmid).</li><br /> | ||
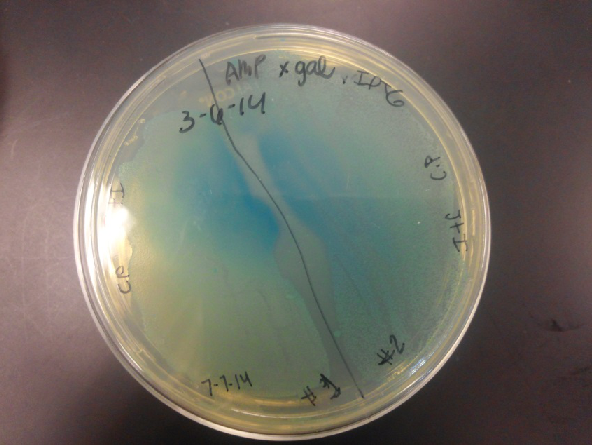
| - | <li>The plates had a 1:7 ratio of white to red colonies. Transformants were selected from TET plates expanded in LB with the appropriate antibiotic, rolled in the 37 Celsius incubator overnight. The next day we plated on TET and 2 % X-gal plates. 2% X-gal plates were made by Kevin using the protocol in PT’s lab binder. X-gal plates contain | + | <li>The plates had a 1:7 ratio of white to red colonies. Transformants were selected from TET plates expanded in LB with the appropriate antibiotic, rolled in the 37 Celsius incubator overnight. The next day we plated on TET and 2 % X-gal plates. 2% X-gal plates were made by Kevin using the protocol in PT’s lab binder. X-gal plates contain galactopyranoside which detects the activity of converting lactose into glucose and galactase. Betagalactosidase activity is present in our K1477014 due to the indication of a blue color on the IPTG and X-gal plates. </li><br /> |
<li>Ten 1.8 mL cryogenic vials with 50/50 of our cells from composite tube #1 and #2 and 40% glycerol were prepared and stored in the -80 freezer as reserve stock.</li><br /> | <li>Ten 1.8 mL cryogenic vials with 50/50 of our cells from composite tube #1 and #2 and 40% glycerol were prepared and stored in the -80 freezer as reserve stock.</li><br /> | ||
<li>Performed an upstream and downstream cut of I712074 (T7 promoter), K112806(Endolysin) with pSB1AT3(destination plasmid). Plated and picked colonies waiting to see results. We named the endolysin part as K1477030. | <li>Performed an upstream and downstream cut of I712074 (T7 promoter), K112806(Endolysin) with pSB1AT3(destination plasmid). Plated and picked colonies waiting to see results. We named the endolysin part as K1477030. | ||
Revision as of 14:10, 15 October 2014
Data Review 10/14/14

- Data from a replication of Stanick’s assay with K1477014: Cells were grown overnight in LB or LB/glucose. Cells in LB were stimulated with 200 uM IPTG for 1 hour and then all cell cultures were divided into 1mL aliquots to which calcium chloride and magnesium sulfate were added. T7 bacteriophage (1 x 10^5 pfu) was added to the appropriate culture and the absorbance at A600 was followed until virus lysis was evident. Cell cultures were then transferred to SpinX filter devices and the media was separated from the cells. Aliquots of filtrates from cells only, T7-treated cultures, or from T7-treated culture were transferred to a microtiter plate. Buffer Z with 2 mercaptoethanol was added and absorbance at 415 nm was determined after 30 minutes.
- Results: Both E.coli C and K1477014 cells stimulated with IPTG “leaked” β-gal activity (Cells only). Top10 cells were negative as expected. β-gal activity was suppressed in all cell lines by glucose as expected. T7 treatment resulted in an increased release of β-gal activity from both E. coli C and K1477014. The combination of T7 lysates from Top10 cells stimulated with IPTG and K1477014 cells grown in glucose did not show any β-gal activity. .
- Conclusion: Cells stimulated with IPTG and subjected to this protocol release β-gal activity. This might be due to β-gal activity that accumulated in the media as the cells grew. Another possibility is that β-gal activity was released when the calcium chloride or magnesium sulfate solutions were added before virus lysis or perhaps released during centrifugation of the cells in the SpinX columns.
- Most important however is that β-gal activity was found in the medium of lysed and unlysed K1477014 suggesting that the LacZ alpha and omega polypeptides stay combined once they are released! That is great news!
- T7 treatment caused an increase in the release of β-gal enzyme from the E. coli C and the β-gal enzyme (alpha-omega polypeptide combo) from K1477014.
- Unfortunately, the combination of T7 lysates of Top 10 cells treated with IPTG, which should have formed LacZ omega polypeptides and the K1477014 cells treated with glucose, which should have formed only LacZ alpha polypeptides, did not have any detectable β-gal activity.
- This lack of activity could be due to the slow association of the alpha and omega fragments, maybe slowed by the Buffer Z and calcium and magnesium salts or, worse, it means that the alpha and omega polypeptides have to associate inside a cell to be joined into a working enzyme. We can study this further by letting the combination of fragments incubate together for hours in hopes that β-gal activity appears. If it does that is great because that is the proof of principle that we are after.
Week of 8/3/14 through 8/9/14
- During this week, the team realized that the contamination was coming from our stock of cells that was being kept in the -80C freezer. We have no idea how they became contaminated, seeing as continued to use the line of cells we froze for a week after we froze them, but before any contamination occurred. Nonetheless, one of our advisers has procured a new stock of top 10 E. coli cells for us to use.
- After unsuccessful runs of electrophoresis gels, we have run out of DNA to try to measure. So, we took some time out this week to streak, grow, and purify lines of our K1477014 and K1477030 parts on both pSB1C3 and pSB1AT3 backbones.
Week of 7/13/14 through 7/19/14
- This week we worked towards switching our part's backbone from pSB1AT3 to pSB1C3. We had some difficulty doing this. The first attempt was fruitless, but on our second attempt we had success and we now have a stock of our part on a chlor-resistant backbone.
- Unfortunately, We no longer have any stock of our original part. So, now we are again following the 3A Biolabs Biobrick Assembly Manual to reconstruct our parts K1477014 and K1477030. Our first try yielded nothing.
- Our second run through of the protocol resulted in the growth of red colonies on chlor plates after electroporation of our part into Top 10 cells. However, we need white colonies to grow because this will point towards the rfp being repressed when the three parts come together. Red colonies are a sign that we did not have a successful assembly of our parts.
Week of 5/25/14 through 5/31/14
- We transformed Top 10 cells through electroporation with registry parts K112806 (Endolysin,) I732020 (LacZoperon Mutant,) J23102 (p-con) and I712074 (T7 promoter.)
- After streaking on plates, and leaving them in the incubator for a day, there was growth on all plates, showing that a successful transformation occured.
- Plates containing J23102 were red-another sign of success. Unfortunately, it looks like contamination has arisen as well, and is spreading on plates with I712074 and K112806 growth.
- The contamination persisted all week, but we were able to get plates with transformed parts that contained no contamination growth.
- We then ran a DNA purification protocol of J23102 and I732020 to get them ready for a biobrick assembly protocol.
 "
"