2D detection of IPTG and HSL
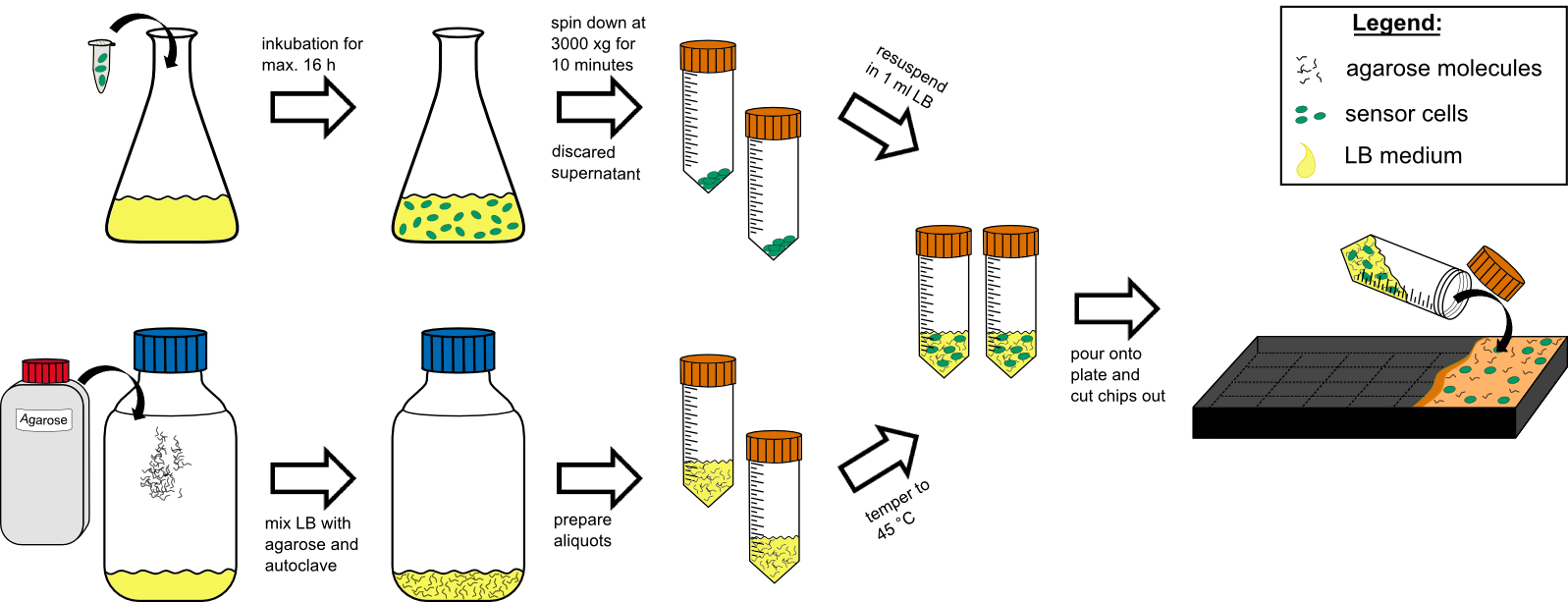
Chip production
Cell preparation
- over night culture of sensor cells (50 mL in a 250 mL flask with) max. 16 h
- centrifuge all 50 mL by 3000 g for 10 min at RT (21 °C).
- discard the supernatant
- re-suspend the pellet in 1 mL tempered (~21 °C) LB-medium .
Agar preparation
- autoclave 50 mL medium with 1.5 % (w/v) agarose (has to be multiplied with the number of chips prepared).
- cool it down to 45 °C in a water bath.
Chip preparation
- mix the cooled medium with the cells by inverting gently.
- pour it in the chip form, avoiding bubble formation (!).
- wait for approximately 20 min until the agar has solidified.
- cut out the chips with a scalpel.
- put two chips into a labeled petri dish and store additional 4 chips in labeled petri dishs in the refrigerator.
- incubate two chips for 1 h at 37 °C prior to induction.
Measurement of fluorescence
Culture medium and culture conditions
Media
LB medium
- weight components
- 5 g/L NaCl
- 10 g/L tryptone
- 5 g/L yeast extract
- (15 g/L agar for plates)
- fill up to 1 L with deionized water
- mix well by shaking
- autoclave
- autoclaving tape, caps slightly unscrewed
- base of the pot has to be covered with deionized water
- close lid
- heat level 3 until the pressure valve opens
- reduce heat level to 1.5
- set timer to 20 minutes
- turn heater off
- wait until the pressure valve retracts (30-45 minutes)
- open, close caps & shake
- for plates, wait until you can touch the bottle (<60 °C, clean bench!)
- add antibiotics (1 µL/mL) and shake (gloves!)
TB medium
- components 1:
- 4 mL/L glycerol
- 12 g/L tryptone
- 24 g/L yeast extract
- fill up to 900 mL with deionized water
- mix well by shaking
- autoclave
- components 2:
- 0.17 M KH2PO4
- 0.72 M K2HPO4
- dissolve in 100 mL deionized water and sterilize it by passing it through a filter
- after autoclaving and cooling down, add sterile phosphate solutions
Hartmans minimal medium (HM)
M9 minimal medium (M9)
| Components for 1 L | Volume
|
| bidest. water | 778.667 mL
|
| 10x Salt solution | 100 mL
|
| Magnesiumsulfatehaptahydrate (10 mM) | 100 mL
|
| Glucose 20% (w/v) | 20 mL
|
| 1000x Trace elements | 1 mL
|
| Thiamin (1 mM) | 0.333 mL
|
| 10x Salt solution
|
| Component' | Final concentration | Concentration in stock solution
|
| BisTris | 95 mM | 200,000 mg/L
|
| Ammmonium chloride | 60 mM | 32,100 mg/L
|
| Sodium citrate | 12.5 mM | 27,000 mg/L
|
| Monopotassium phosphate | 3 mM | 4,170 mg/L
|
| Dipotassium phosphate | 0.7 mM | 1,590 mg/L
|
| 1000x Trace elements
|
| Component | Final concentration | Concentration in stock solution
|
| Iron(III) chloride | 50 mM | 13,515 mg/L
|
| Calcium chloride | 20 mM | 2,220 mg/L
|
| Manganese(II) chloride | 10 mM | 1,258 mg/L
|
| Zinc sulfate | 10 mM | 1,615 mg/L
|
| Cobalt(II) chloride | 2 mM | 260 mg/L
|
| Copper(II) chloride | 2 mM | 269 mg/L
|
| Nickel(II) chloride | 2 mM | 259 mg/L
|
| Sodium molybdate | 2 mM | 412 mg/L
|
| Sodium selenite | 2 mM | 346 mg/L
|
| Boric acid | 2 mM | 124 mg/L
|
| Hydrochloric acid | 1 mM | 20 mL
|
SOC
- components
- 0,5 % yeast extract
- 2 % tryptone
- 10 mM NaCl
- 2.5 mM KCl
- 20 mM MgSO4
- fill up with deionized water
- adjust to pH 7.5 with NaOH
- after autoclaving, add 20 mM sterile glucose solution (filter sterilization)
Genetic methods
Cloning
Restriction Digest
Ligation
Gibson Assembly
The Gibson Assembly was conducted according to the protocol published by New England Biolabs.
- Set up the reaction according to the table below on ice (2-3 fragment assembly).
- Incubate samples in a thermocycler at 50°C for 15 minutes when 2 or 3 fragments are being assembled or 60 minutes when 4-6 fragments are being assembled. Following incubation, store samples on ice or at –20°C for subsequent transformation.
- Transform NEB 5-alpha Competent E. coli cells with 2 μl of the assembly reaction, following the transformation protocol.
| Total Amount of Fragments | 0.02-0.5 pmols
|
| Gibson Assembly Master Mix (2X) | 10 µl
|
| Deionized H2O | 10-X µl
|
| Total Volume | 20 µl
|
Transformation
Heat Shock
- thaw cells on ice
- add 1 µL of plasmid DNA
- incubate on ice for 30 min
- heat shock at 42 °C for 60 s
- incubate on ice for 5 min
- add 200 µL of SOC media
- incubate at 37 °C for 2 h
- plate 20 and 200 µL on plates supplemented with the appropiate antibiotic
Electroporation
- add 1 μL plasmid to electrocompetent cells
- put DNA/ cell suspension in electroporation cuvette
- wipe dry the electroporator
- use a small plastic pipette to place the cells
- pulse: 2.5 kV, 200-400 Ω, 25 μF (for E.coli)
- immediatly add 1 mL LB and incubate for 2 h at 37 °C
- plate 50 μL on selective medium plate
- centrifuge the rest (3000 g, 20 min), discard supernatant, re-suspend the pellet in 50 μL LB and plate it on selective medium plate
PCR
We have used several different types of PCR throughout our project:
- colony PCR
- check PCR
- gradient PCR
- SOE PCR
- touchdown PCR
- QuikChange(Ligation-During-Amplification)
The scope of appplication as well as the conduct are described below.
Colony PCR /Check PCR
With GoTaq Mast Mix
- 12.5 µl GoTaq Master Mix
- 1 µl primer_F
- 1 µl primer_R
- pick colony with tip and suspend in PCR tube
- 9.5 µl ddH2O
| parameter | duration | temp [°C] |
|
| denature | 5:00 | 95 |
|
| anneal | 00:30 | 56 | 30 cycles
|
| elongate | 01:00 per kb | 72
|
| denature | 00:30 | 95
|
| elongate | 05:00 | 72 |
|
| store | forever | 8
|
gradient PCR
SOE PCR
touchdown PCR
QuikChange
Analytical methods
Agarose gel electrophoresis
Separation of DNA or RNA
- take 5µl of the PCR product
- mix with 1µl loading dye
- apply onto agarose gel together with a marker
- run at 120°C for 40 minutes for a full gel
SDS-PAGE
Cell preparation
- lysis cell pellet in lysis buffer
- centrifuge for 15 min at 13.000 rpm
- mix the supernatant with 2x lammli buffer with β-mercaptoethanol
- denatured for 5 min at 95 °C
- sample to the gel
For some SDS-PAGEs, we used BioRad ready made gels.
The recipe of the self-made SDS is as follows:
1.5x Buffer
- 1.5 M Tris-Cl pH = 8.8
- in 1 L is 40 ml 10 % SDS
Gels
|
| 0.75 mm 12 % RUNNING Gel
| 1 mm 4 % STACKING Gel
|
|
| 1x | 2x | 4x
| 1x | 2x | 4x
|
| H2O
| 1.65 mL | 3.3 mL | 6.6 mL
| 1.5 mL | 3 mL | 6 mL
|
| 1.5x Gel Buffer
| 1.3 mL | 2.6 mL | 5.2 mL
| 0.65 mL | 1.3 mL | 2.6 mL
|
| 30 % Acrylamide (37.5:1)
| 2 mL | 4 mL | 8 mL
| 0.325 mL | 0.65 mL | 1.3 mL
|
| 10 % APS
| 50 µL | 100 µL | 200 µL
| 25 µL | 50 µL | 100 µL
|
| TEMED
| 10 µL | 20 µL | 40 µL
| 5 µL | 10 µL | 20 µL
|
Run gel
- apply the prepared samples together with a protein marker on the gel
- run the gel for 10 min at 60 V and after that for ca. 60 min at 120 V
Bradford assay
Determination of protein concentration
Measurement of fluorescence
The measurement of fluorescence was performed using the Synergy Mx (BioTek) microplate reader and the Gen5 software.
- volume of sample in each well: 100µl
- measure GFP fluorescence at an excitation wavelength of 496 nm and an emission wavelength at 516 nm
Measurement of optical density
Depending on the number of samples, two different devices were used for measurement of optical density, the Unico Spectrophotometer 1201 (Fisher Bioblock Scientific) and the Synergy Mx (BioTek) microplate reader.
|
 "
"