Team:SUSTC-Shenzhen/Notebook/Construction of gRNA for HIV or HBV carried plasmids
From 2014.igem.org
Notebook
Elements of the endeavor.
Contents |
Construction of plasmids carrying the gRNA for HIV/HBV
2014/10/6~2014/10/10
Construction of plasmids carrying the gRNA for HIV/HBV
Materials
Enzyme: BbsI
Buffer: Buffer2.1
T4 DNA ligase, T4 DNA ligase buffer
The usual things needed for transformation
Methods
Restriction digestion (with BbsI) for gRNA-carried plasmid backbones (with2X/5X/7XUAS sequences) & gRNA for HIV/HBV DNA sequence
Digestion system(unit:ul):
| DNA | BbsI | Buffer | Total | ||
|---|---|---|---|---|---|
| Vector | gRNA-carried plasmid backbones (with2X/5X/7XUAS sequences) | 1 | 1 | 1 | 10 |
| Insert | gRNA for HIV/HBV DNA sequences | 8.5 | 0.5 | 1 | 10 |
Incubate at 37C for 4 hours.
Electrophoresis and Agarose gel DNA purification
We perform electrophoresis and Agarose gel DNA purification for the digested gRNA-carried plasmid backbone to acquire the vector for the following ligation step.
ligation for gRNA-carried plasmid backbones and gRNA for HIV/HBV DNA sequences
Ligation system [unit: ul]:
| Vector | Insert (48ng/ul) | Vector(needed 25ng) | Insert(needed 2.8ng) | T4 ligase | Buffer | H2O | Total |
|---|---|---|---|---|---|---|---|
| 2X (15.7ng/ul) | HIV | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 2X (15.7ng/ul) | HBV_1 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 2X (15.7ng/ul) | HBV_2 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 5X(15.8ng/ul) | HIV | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 5X(15.8ng/ul) | HBV_1 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 5X(15.8ng/ul) | HBV_2 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 7X (15.4ng/ul) | HIV | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 7X (15.4ng/ul) | HBV_1 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
| 7X (15.4ng/ul) | HBV_2 | 4.0 | 0.06 | 1 | 2 | 12.9 | 20 |
Protocol
a. incubate at RT for 15min
b. heat-inactivation at 65C for 10min
d. Transformation
Performed as protocol…
5ul ligation reaction to 50ul competent cell
Results
Luckily, each of our plates has several colonies grown out, which meant that our first step for the construction of gRNAforHIV/HBV-carried plasmids succeeded!
Thus, we picked up one monoclone for each type of gRNAforHIV/HBV-carried plasmids, incubate in shaker, 200rpm, 37°C for 14 hours.
Plasmid purification for gRNAforHIV/HBV-carried plasmids (with 2X/5X/7X) plasmids
Materials
TIANprep Plasmid Kit
Methods
Performed as the usual protocol provided…
Verification with AgeI & BbsI
Materials
Enzyme:BbsI, AgeI
Buffer: Buffer2.1, Cutsmart
Methods
Digestion system: [unit: ul]
| DNA | AgeI | BbsI | Buffer | H2O | Total |
|---|---|---|---|---|---|
| 0.5 | 0.5 | 0.5 | 1 | 7.5 | 10 |
Incubate at 37C, 4hours
Results
Assumption< br> Successful situation:< br> The DNA fragment inside the two BbsI sites has been replaced by a gRNA for HIV/HBV DNA sequence. So, there is no BbsI site on each plasmid now.< br> Plasmid just became linearized when digested by both A & B. < br>
Failed situation:
The DNA fragment inside the two BbsI sites hasn’t been replaced. So, there are still BbsI sites on each plasmid now.
Plasmid will be cut into two fragments when digested by both A & B.
Actual results
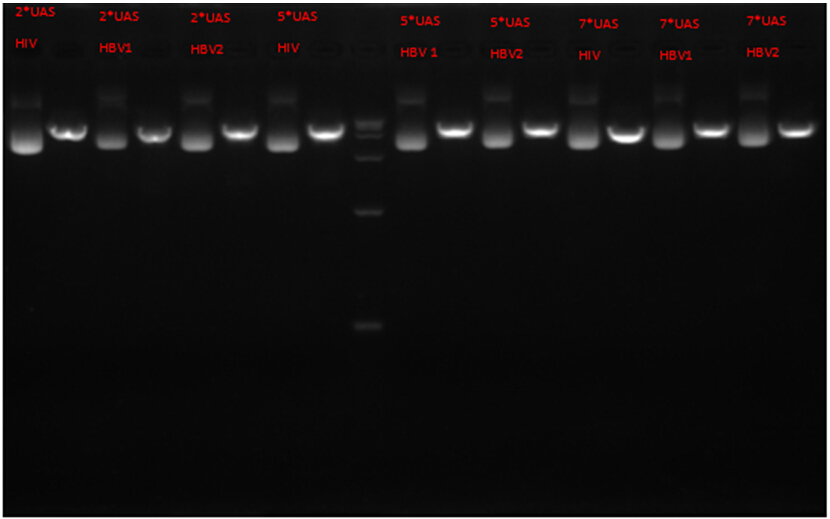
Obviously, all of the nine plasmids matched the successful items above.
Thus, we can pronounce that our construction of gRNAforHIV/HBV carried plasmids has succeeded!
Plasmid purification (no endotoxin) for gRNAforHIV/HBV carried plasmids
Since we have confirmed that all of the nine gRNAforHIV/HBV carried plasmids we constructed are successful, we performed plasmid purification (no endotoxin) for them.
Materials
EndoFree Plasmid Kit
Methods
Performed as the usual protocol provided…
 "
"