Team:SUSTC-Shenzhen/Notebook/CRISPR/Point-mutation-PCR-for-mCherry-with-BbsI-site-mutation
From 2014.igem.org
Notebook
Elements of the endeavor.
Contents |
Point-mutation PCR for mCherry (with BbsI site mutation)
2014/8/21 To eliminate the BbsI site on mCherry fragment.Materials
Q5TM High-Fidelity 2X Master Mix (DNA polymerase, dNTPs, Mg++, buffer)
mCherry forward primer(50p mol/ μl)
mCherry reverse primer with 2*UAS (50p mol/ μl)
Pointmutation Primer F&R (50p mol/ μl)
Template DNA (pBX-084 PB5 - HS4 - TRE - mCherrynuc - 2A - BlaloxN - GpA - HS4-PB3)
dd H2O
Methods
Point-mutation PCR for mCherry
1.Add 1μl template DNA into 9μl dd H2O to dilute the template DNA.
2.Add 1μl pointmutation primer forward and 1μl pointmutation primer reverse into 18μl dd H2O to dilute the pointmutation primers.
3. The total volume is 125μl, including 62.5 μl Q5TM High-Fidelity 2X Master Mix (dilute it from 2X to 1X), 2.5μl mCherry forward primer, 2.5μl mCherry reverse primer with 2*UAS, 1 μl diluted template DNA , 0.5μl diluted pointmutation primer F&R and 56μl dd H2O. And divide the 125 μl solution into 5 PCR tubes in average. Attention, the operation should be finished on ice.
4. Set the PCR machine and run:
Stage1: Initial denaturation: 98°C,30s.
Stage2: Denaturation :98°C,10s
Anneal: 52°C,55°C,58°C,61°C,64°C,respectively.20s.
Extension: 72°C, 30s.
35 cycles.
Stage3: Final extension:72°C, 5 min.
Hold: 4°C.
Electrophoresis for the product of PCR
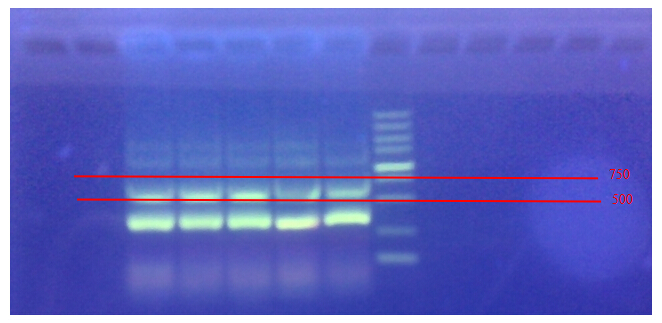
The band between 500 and 750 is what we want. According to gel eletrophoresis, the most suitable anneal temperatures for these primers are 55°C and 58°C .
Gel extraction
Tiangen Miniprep gel extraction kit (50 rxn) As the protocol Nanodrop test :
| number | 1 | 2 |
|---|---|---|
| Conc (ng/ul | 59.3 | 37.4 |
Verification by enzyme digestion
BbsI digestion(14:46~18:46)
| Component | Volume |
|---|---|
| BbsI | 0.5ul |
| DNA | 2ul |
| 10X NEBuffer 2.1 | 1ul |
| ddH2O | 6.5ul |
| Total | 10ul |
BspEI digestion(14:46~18:46)
| Component | Volume |
|---|---|
| BspEI | 0.5ul |
| DNA | 2ul |
| 10X NEBuffer 3.1 | 1ul |
| ddH2O | 6.5ul |
| Total | 10ul |
BbsI and BspEI digestion(14:46~18:46)
| Component | Volume |
|---|---|
| BbsI | 0.8ul |
| BspEI | 0.2ul |
| DNA | 2ul |
| 10X NEBuffer 3.1 | 1ul |
| ddH2O | 6ul |
| Total | 10ul |
Gel electrophoresis
Making the agarose gel (1%)
| Component | Volume |
|---|---|
| TAE | 50ml |
| Agarose | 0.5g |
| Gene Green | 1ul |
Results of electrophoresis
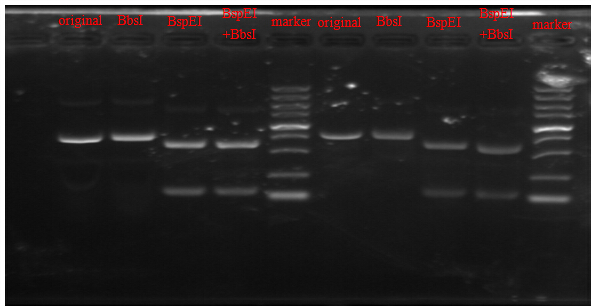
From the picture, we can see that, the figure of plasmid digested with BbsI was the same as that of the original plasmid without digestion, indicating that the BbsI site have been successfully removed from the mCherry fragment by our Point-mutation PCR. Further evidence for successful mutation was provided by the results of digestion with BbsI& BspEI simultaneously and with BspEI only. Since the profile of the plasmid digested by both BbsI and BspEI was exactly the same as the figure of digestion with BspEI singly, we can concluded that the BbsI site has been mutated out on the mCherry fragment.
 "
"