Team:Groningen:Project:Toolbox
From 2014.igem.org
Project
>
Toolbox
Lactococcus lactis toolbox
We started with a toolbox that will make it possible to use
Lactococcus lactis as a chassis. This toolbox contains a vector,
a set of constitutive promoters, several genes from the nisin operon
and an excellent fluorescent marker.
Nisin operon
An important part of our project, that will also form a nice addition
to the toolbox, is the nisin operon. The nisin operon is responsible for
making and secreting the lantibiotic nisin in certain L. lactis
species. Nisin inhibits the growth of a broad range of Gram positive
bacteria, of which many are spoilage bacteria or pathogens. Nisin
is therefore extensively used in the food industry as a preservative.
Nisin forms pores in the membrane of the bacteria it kills and inhibits
the peptidoglycan synthesis.2
The operon consists of the genes NisA, NisB, NisT, NisC, NisI,
NisP, NisR, NisK, NisF, NisE and NisG and their related promoters, see
figure 1. NisA is the gene that codes for the nisin precursor (1), that will
be modified and transported out of the cell. The nisin precursor its
serines and threonines are dehydrated by NisB and then the precursor
is cyclized by NisC. After this process, the precursor is transported out
of the cell (2). Here, the lead peptide is cut off by NisP (3).
Additional genes of the nisin operon are NisR and NisK (4). NisK is a membrane receptor that activates NisR when it binds nisin. NisR can then activate nisin inducible promoters, like PNisA and PNisI. At last, there are the nisin immunity genes, NisI, NisF, NisE and NisG (5). NisI, a lipoprotein, serves as the first defense, and is therefore always present in the cell that has nisin immunity. NisF, NisE and NisG form an ABC-exporter together. They provide extra immunity and become present in the cell when nisin is encountered in the environment.3
Additional genes of the nisin operon are NisR and NisK (4). NisK is a membrane receptor that activates NisR when it binds nisin. NisR can then activate nisin inducible promoters, like PNisA and PNisI. At last, there are the nisin immunity genes, NisI, NisF, NisE and NisG (5). NisI, a lipoprotein, serves as the first defense, and is therefore always present in the cell that has nisin immunity. NisF, NisE and NisG form an ABC-exporter together. They provide extra immunity and become present in the cell when nisin is encountered in the environment.3
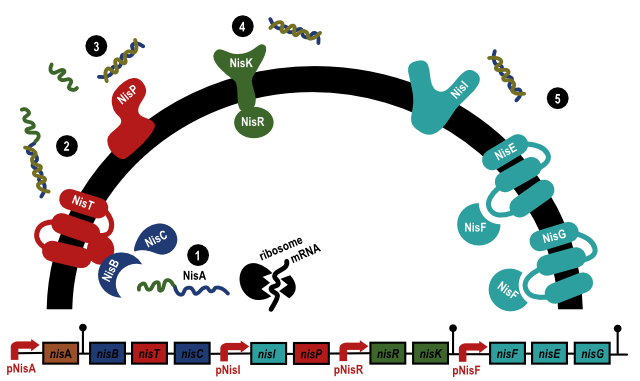
Figure 1:
Function of the different genes of the nisin operon.
Of the nisin operon, NisA, NisC, NisR, NisK and PNisA were sent to
iGEM HQ. NisA can be used in L. lactis NZ9800, a NisA deficient strain, to let it produce
nisin.4 PNisA is a promoter that is induced by nisin. NisR and NisK can
be used to detect nisin and activate nisin inducible promoters.

Vector
To get BioBricks in L. lactis, a compatible backbone is needed. For this,
we used a well known plasmid that is known to work in L. lactis, pIL253,
and made this BioBrick compatible. The plasmid is a shuttle vector that can be
used in L. lactis and Bacillus subtilis. It carries erythromycin
resistance as a selection marker and has a mRFP coding device between the
BioBrick prefix and suffix, so selection on inserts in this vector is
possible.
Promoters
For expressing genes in L. lactis, promoters are needed.
Together with the most standard RBS and the standard double terminator they get genes transcribed and translated. The set of promoters that is used for the L. lactis toolbox is the
promoter collection that was BioBricked by the Uppsala iGEM team in 2013. The set
consists of the promoters CP1, CP8, CP11, CP29,
CP30, CP41 and CP44.
The promoters originate from L. lactis and have first been described
by Jensen and Hammer in 1998.1
The promoters were placed in front of a fluorescent marker in L. lactis,
so their relative strength is known.
Fluorescent marker
Last but not least, a fluorescent marker that has a high performance in
L. lactis was BioBricked and added to the toolbox. This marker
is a version of the sfGFP protein, called sfGFP(Bs), that was optimized for Bacillus subtilis.
When the optimized sfGFP was tested, it showed high performance in
L. lactis.5 The sfGFP(Bs) can be used for the testing of
constructs, quantification of promoters or it can be coupled to other
proteins to track them inside or outside the cell.

Figure 2:
E. coli expressing sfGFP(Bs), combined with the promoters CP41 and CP44.
References
1. Jensen, P.R. and Hammer, K. (1998) Artificial promoters for metabolic optimization.
Biotechnol. Bioeng. 58: 191-195
2. Zhou, H. et al. (2014) Mechanisms of nisin resistance in Gram-positive
bacteria. Ann. Microbiol. 64: 413-420
3. Cheigh, C. I. and Pyun, Y.R. (2005) Nisin biosynthesis and its
properties. Biotechnol. Lett. 27: 1641-1648
4. Kuiper, O.P. et al. (1993) Characterization of the nisin gene-cluster NisABTCIPR of Lactococcus lactis -
requirement of expression of the NisA and NisI genes for development of
immunity. Eu. J. Biochem. 216: 281-291
5. Overkamp, W. et al. (2013) Benchmarking various green fluorescent
protein variants in Bacillus subtilis, Streptococcus pneumoniae, and
Lactococcus lactis for live cell imaging. Appl. Environ. Microbiol. 79: 6481-6490
 "
"

