Team:BUCT-China/small-dialogpj4
From 2014.igem.org

PROJECT
Biosensors
LuxAB---comprehensive biosensor
Luminescent Bacteria Method:
Adoption of luminescent bacteria for air pollution monitoring, antibiotics screening and rapid detection of water attracts wide attention in recent years. Luminescent bacteria is a kind of bacteria that can produce visible fluorescence in normal conditions, while its bioluminescence is produced by molecular oxygen and catalyzed by luciferase inside the cell. Luciferase oxidates flavin mononucleotide (FMNH2) and long chain fatty aldehyde in the reduction state to FMN and long chain fatty acid, at the same time emits blue-green light in 450 ~ 490 nm. Equation can be expressed as follows:
FMN+NAD(P)H-→FMNH2+NAD(P)
FMNH2+RCHO+O2-→FMN+RCOOH+H2O+hv
2、 the effects of Web site, poster and other artists are not so beautiful;The process of luminescence involves a variety of substances, which are products of bacterial metabolism. When external condition worsens, its luminous way will be affected in a very short period of time, resulting in suppression or shutdown of luminescent reaction. As a result, luminescence of bacteria is positively correlated to the concentration of toxin in condition, what’s more, the correlation can be approached by a linear function. Based on the mechanism, luminescent bacteria have been widely adopted as a biological indicator in rapid quantification of toxin in the environment.
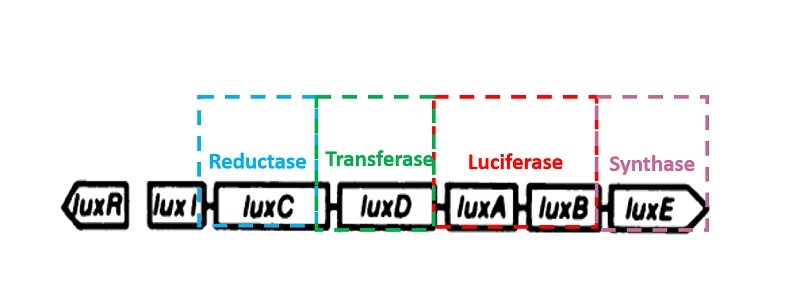
Luminescent bacteria luciferase gene
Bacteria luminescence is a consequence of its luciferase catalytic metabolic process. The luciferase encoding gene is LuxAB. All genus of luminescent bacteria have five core constitute lux gene operon, order of which is luxCDABE. Bacteria luciferase is composed of two subunits heterodimer, including luciferase alpha and beta subunits coded by luxA and luxB respectively. LuxC, luxD, luxE encode for reduction of fatty acid synthesis fatty aldehyde enzyme complexes of three subunits: reductase, transferase, synthetase, respectively. Mancin (1988) successfully cloned the photobacterium phosphoreum’s luciferase operon luxCDABFE and expressed it in Escherichia coli. Except for methods though plasmid, lux gene can also be integrated into the cell chromosome by genetic engineering techniques in pursuit of stability. What’s more, bioluminescent detection through lux gene is easy to achieve, and it is fast, simple and sensitive. Hence the bioluminescent detection method has extensive research and application in gene transduction, expression and regulation, environmental chemistry and its toxicity detection,etc.
Reference:
Shi J, Frymier P D. Toxicity of metals and organic chemicals evaluated with bioluminescence assays[J]. Chemosphere, 2005, 58: 543-550.
Ru CX, Yang SD. Luminescent bacteria lux report gene system evaluation and application [J]. Microbiology, 2000, 27(4):297-299.
MerR ---Hg(II) Biosensor
Metals can pose a particular problem to bacteria,because essential metals may be limiting in the environment, requiring active uptake mechanisms to import minimum concentrations of essential metals (Butler 1998; Outten et al. 2000).Bacteria can encounter purely toxic metals, such as Hg, Cd and As, which have no beneficial role in cellular metabolism, and must be avoided, removed or neutralized metal. The key first step in how bacteria respond to varying levels of both toxic and essential metals in the internal environment of the cell is due to the metal sensor and regulator proteins that they encode. There are several known types of prokaryotic metal ion sensing regulators. These include the MerR and Fur families of regulators, the ArsR/SmtB family repressors and several other structural regulator types .
MerR – a mercury sensing gene activator
A key class of prokaryotic metal ion responsive activators is the MerR family, of which the mercuric ion sensing MerR is the archetype. The closely-related 144 amino-acid MerR proteins from the mercuric ion resistance (mer) operons from transposons Tn501 and Tn21 have been the most heavily studied of these proteins (reviewed in Summers 1992, 1986; Hobman & Brown 1997;Outten et al. 2000a; Barkay et al. 2003; Brown et al.2003, and references therein), and the mechanism of Hg(II) resistance is now well known (reviewed most recently by Barkay et al. (2003)).
In the mer operon from Tn501, MerR binds to operator DNA within a divergent promoter and regulates both its own expression and expression of the downstream structural genes. The MerR family of metal-binding, metal-responsive proteins is unique in that they activate transcription from unusual promoters and coordinate metals through cysteine residues. They have conserved primary structures yet can effectively discriminate metals in vivo.
Activation of transcription from the PmerT promoter by MerR in response to Hg(II) is hypersensitive (Ralston &O’Halloran 1990; Rouch et al. 1995), with virtually total induction of promoter activity occurring across a very narrow Hg(II) concentration rang. MerR has a strong selectivity for Hg(II) (Ralston & O’Halloran 1990)and Hg(II) has a very high affinity for MerR (Shewchuk et al. 1989b).
The model for regulation of the mer promoter
The current model for MerR activation of transcription proposes that expression of MerR (in the absence of MerR and Hg (II) in the cell) proceeds from the PmerR promoter. In the absence of Hg(II), the MerR homodimer binds to the operator region within the divergent promoter with binding centered on the dyad symmetrical DNA sequence between the -35 and -10 sequences of PmerT, slightly repressing transcription of the structural gene promoter, and repressing transcription of merR from the PmerR promoter. This is probably due to MerR interfering with RNA polymerase (RNAP) binding, or open complex formation. PmerT is unusual, it has a 19 bp spacing between the -35 and -10 sites , rather than the 16–18 bp spacing found in most prokaryotic promoters (Harley & Reynolds 1987). This makes PmerT suboptimal for RNAP recognition of, and binding to, the -35 and -10 sequences, preventing formation of the open complex and transcriptional activation (see Browning & Busby 2004). In the absence of Hg(II), the tight binding of the apo-MerR homodimer to PmerT causes a further bending of the promoter DNA to itself, making it even less ideal for RNAP binding (Ansari et al. 1995). Once the MerR homodimer has bound to merOP, recruitment of RNAP to the mer promoter occurs (Heltzel et al. 1990), and MerR has been shown to cross-link to several subunits of RNAP (Kulkarni & Summers 1999). In the absence of Hg(II) the ternary complex of MerR, RNAP, and merOP represses transcription of the mer structural genes. In the presence of mercuric ions, one Hg(II) per MerR homodimer (O’Halloran et al. 1989; Shewchuk et al. 1989a) coordinates in a trigonal manner to three essential cysteine residues of the MerR homodimer, two cysteines from one monomer, and one from the other (Helmann et al. 1990; Utschig et al. 1995). Hg(II) binding to the MerR homodimer results in both a relaxation of the DNA bends induced by apo-MerR, and both DNA distortion (Frantz &O’Halloran 1990) and an allosteric under winding of the promoter sequence by approximately 33(Ansari et al. 1992). The underwinding of the promoter DNA aligns the-10 and -35 sequences, such thatRNApolymerase can recognize and bind to these sites, initiating transcription from PmerT (Heltzel et al. 1990; Ansari et al.1992, 1995).

Reference:
Butler A. 1998 Acquisition and utilization of transition meal ions by marine organisms. Science 281, 207–210.
Outten FW, Outten CE, O’Halloran TV. 2000a Metalloregulatory systems at the interface between bacterial metal homeostasis and resistance. In:
Storz G, Hengge-Aronis R,(eds.) Bacterial stress responses. American Society of Microbiology;Washington DC: pp. 145–157.
Outten FW, Outten CE, Hale J, O’Halloran TV. 2000b Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue CueR. J Biol Chem 275, 31024–31029
Barkay T, Miller SM, Summers AO. 2003 Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev 27, 355–384.
Hobman JL, Brown NL. 1997 Bacterial mercury resistance genes. In: Sigel A, Sigel H, eds. Metal Ions in Biological Systems. New York: Marcel Dekker; 34, 527–568.
Brown NL, Stoyanov JS, Kidd SP, Hobman JL. 2003 The MerR family of transcriptional regulators. FEMS Microbiol Rev 27, 145–163.
O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG.1989 The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex.Cell 56, 119–129.
Rouch DA, Parkhill J, Brown NL. 1995 Induction of bacterial mercury- and copper-responsive promoters: functional differences between inducible systems and implications for their use in gene-fusions for in vivo metal biosensors. J Ind Microbiol 14, 249–253.
Shewchuk LM, Verdine GL, Walsh CT. 1989b Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities.Biochemistry 28, 2331–2339.
Harley CB, Reynolds RP. 1987 Analysis of Escherichia coli promoter sequences. Nuc Acids Res 15, 2343–2361.
Browning DF, Busby SJW. 2004 The regulation of bacterial transcription initiation. Nat Rev Microbiol 2, 57–65.
Ansari AZ, Bradner JE, O’Halloran TV. 1995 DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374, 371–375.
Kulkarni R, Summers AO. 1999 MerR crosslinks to the a, b and r70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter. Biochemistry 38, 3362–3368.
Utschig LM, Bryson JW, O’Halloran TV. 1995 Mercury-199 NMR of the metal receptor site in MerR and its protein-DNA complex. Science 268, 380–385.
Ansari AZ, Chael ML, O’Halloran TV. 1994 Allosteric under winding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature 355, 87–89.
 "
"