Team:Saarland/Parts
From 2014.igem.org
| (7 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
<br> | <br> | ||
<h1>Parts:</h1> | <h1>Parts:</h1> | ||
| - | |||
<h3>Hyaluronic acid production</h3> | <h3>Hyaluronic acid production</h3> | ||
| - | |||
| - | |||
<h4>Number: [http://parts.igem.org/Part:BBa_K1469001 ''BBa_K1469001'']</h4> | <h4>Number: [http://parts.igem.org/Part:BBa_K1469001 ''BBa_K1469001'']</h4> | ||
| Line 17: | Line 14: | ||
<b>Description:</b><br> | <b>Description:</b><br> | ||
| - | Hyaluronan synthase 2 gene of<i> Heterocephalus glaber</i> with codon adaption for expression in<i> Bacillus megaterium</i>. The enzyme catalyses the linkage of UDP-D-glucuronic acid and UDP-N-acetyl-D-glucosamine to the linear high molecular mass hyaluronic acid chain and is also involved in its secretion.<br> | + | Hyaluronan synthase 2 gene of<i> Heterocephalus glaber</i> with codon adaption for expression in<i> Bacillus megaterium</i>. The enzyme catalyses the linkage of UDP-D-glucuronic acid and UDP-N-acetyl-D-glucosamine to the linear high molecular mass hyaluronic acid chain and is also involved in its secretion.<br><br> |
<html> | <html> | ||
<a href="https://2014.igem.org/Team:Saarland/Modeling"> | <a href="https://2014.igem.org/Team:Saarland/Modeling"> | ||
| Line 25: | Line 22: | ||
</html> | </html> | ||
<br> | <br> | ||
| - | + | <br> | |
| - | <b>Sequence:</b | + | <b>Sequence:</b> |
<!-- EHER LINK ZU [http://parts.igem.org/Part:BBa_K1469001] --> | <!-- EHER LINK ZU [http://parts.igem.org/Part:BBa_K1469001] --> | ||
| - | |||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 64: | Line 60: | ||
<h3>Pathway engineering for optimisation of hyaluronic acid production</h3> | <h3>Pathway engineering for optimisation of hyaluronic acid production</h3> | ||
| - | |||
| - | |||
<h4>Number: [http://parts.igem.org/Part:BBa_K1469002 ''BBa_K1469002'']</h4> | <h4>Number: [http://parts.igem.org/Part:BBa_K1469002 ''BBa_K1469002'']</h4> | ||
| Line 75: | Line 69: | ||
<br> | <br> | ||
| - | <b>Sequence:</b> <br> | + | <b>Sequence:</b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 102: | Line 95: | ||
<a href="https://2014.igem.org/Team:Saarland/4_step#WesternBlot"> | <a href="https://2014.igem.org/Team:Saarland/4_step#WesternBlot"> | ||
<img src="https://static.igem.org/mediawiki/2014/d/d2/Links_Pfeil.1.png" width="40px"> | <img src="https://static.igem.org/mediawiki/2014/d/d2/Links_Pfeil.1.png" width="40px"> | ||
| - | <b> SDS | + | <b> SDS PAGE</b> |
</a> | </a> | ||
</html> | </html> | ||
| Line 116: | Line 109: | ||
Glucokinase of <i>Bacillus megaterium</i>. The enzyme catalyses the phosphorylation reaction of glucose to glucose 6-phospahate.<br> | Glucokinase of <i>Bacillus megaterium</i>. The enzyme catalyses the phosphorylation reaction of glucose to glucose 6-phospahate.<br> | ||
| - | <b>Sequence: </b> <br> | + | <b>Sequence: </b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 149: | Line 141: | ||
Beta-phosphoglucomutase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 6-phospahate to glucose 1-phosphate.<br> | Beta-phosphoglucomutase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 6-phospahate to glucose 1-phosphate.<br> | ||
| - | <b>Sequence:</b> | + | <b>Sequence:</b><br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 179: | Line 170: | ||
UTP--glucose-1-phosphate uridylyltransferase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 1-phospahate to UDP-glucose 1-phosphate.<br> | UTP--glucose-1-phosphate uridylyltransferase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 1-phospahate to UDP-glucose 1-phosphate.<br> | ||
| - | <b>Sequence:</b> | + | <b>Sequence:</b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 211: | Line 201: | ||
Glucose-6-phosphate isomerase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 6-phospahate to fructose 6-phosphate.<br> | Glucose-6-phosphate isomerase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucose 6-phospahate to fructose 6-phosphate.<br> | ||
| - | <b>Sequence:</b> <br> | + | <b>Sequence:</b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 248: | Line 237: | ||
Glucosamine--fructose-6-phosphate aminotransferase (isomerising) of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of fructose 6-phosphate to glucosamine 6-phosphate.<br> | Glucosamine--fructose-6-phosphate aminotransferase (isomerising) of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of fructose 6-phosphate to glucosamine 6-phosphate.<br> | ||
| - | <b>Sequence:</b> <br> | + | <b>Sequence:</b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 290: | Line 278: | ||
Phosphoglucosamine mutase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucosamine 6-phosphate to glucosamine 1-phosphate.<br> | Phosphoglucosamine mutase of <i>Bacillus megaterium</i>. The enzyme catalyses the conversion of glucosamine 6-phosphate to glucosamine 1-phosphate.<br> | ||
| - | <b>Sequence:</b> <br> | + | <b>Sequence:</b><br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
| Line 327: | Line 314: | ||
UDP-N-acetylglucosamine pyrophosphorylase of <i>Bacillus megaterium</i>. The enzyme is bifunctional and catalyses the conversion of glucosamine 1-phosphate to N-acetyl glucosamine 1-phosphate as well as the conversion of N-acetyl glucosamine 1-phosphate to UDP-N-acetyl glucosamine, a key precursor molecule for hyaluronic acid production.<br> | UDP-N-acetylglucosamine pyrophosphorylase of <i>Bacillus megaterium</i>. The enzyme is bifunctional and catalyses the conversion of glucosamine 1-phosphate to N-acetyl glucosamine 1-phosphate as well as the conversion of N-acetyl glucosamine 1-phosphate to UDP-N-acetyl glucosamine, a key precursor molecule for hyaluronic acid production.<br> | ||
| - | <b>Sequence:</b> <br> | + | <b>Sequence:</b> <br> |
| - | + | ||
<html> | <html> | ||
<span style="font-family:Courier New"> | <span style="font-family:Courier New"> | ||
Latest revision as of 12:14, 17 October 2014
Parts:
Hyaluronic acid production
Number: BBa_K1469001
Description:
Hyaluronan synthase 2 gene of Heterocephalus glaber with codon adaption for expression in Bacillus megaterium. The enzyme catalyses the linkage of UDP-D-glucuronic acid and UDP-N-acetyl-D-glucosamine to the linear high molecular mass hyaluronic acid chain and is also involved in its secretion.
 Modelling
Modelling
Sequence:
ATGCACTGTGAACGTTTCTTATGTATTTTACGTATTATTGGTACAACATTATTCGGTGTATCTTTATTATTAGGTATTACAGCTGCTTACATTGTAGGTTACCAA
TTCATTCAAACAGATAACTACTACTTCTCTTTCGGTTTATACGGTGCTTTCTTAGCTTCTCACTTAATTATTCAATCTTTATTCGCTTTCTTAGAACACCGTAAA
ATGAAAAAATCTTTAGAAACACCAATTAAATTAAACAAAACAGTAGCTTTATGTATTGCTGCTTACCAAGAAGATCCAGATTACTTACGTAAATGTTTACAATCT
GTAAAACGTTTAACATACCCAGGTATTAAAGTAGTAATGGTAATTGATGGTAACTCTGATGATGATTTATACATGATGGATATTTTCTCTGAAGTAATGGGTCGT
GATAAATCTGCTACATACATTTGGAAAAACAACTTCCACGAAAAAGGTCCAGGTGAAACAGATGAATCTCACAAAGAATCTTCTCAACACGTAACACAATTAGTA
TTATCTTCTAAATCTGTATGTATTATGCAAAAATGGGGTGGTAAACGTGAAGTAATGTACACAGCGTTTCGTGCTCTCGGTCGTTCTGTTGATTACGTACAAGTA
TGTGATTCTGATACAATGTTAGATCCAGCTTCTTCTGTAGAAATGGTAAAAGTATTAGAAGAAGATCCAATGGTAGGTGGTGTAGGTGGTGATGTACAAATTTTA
AACAAATACGATTCTTGGATTTCTTTCTTATCTTCTGTACGTTACTGGATGGCTTTCAACATTGAACGTGCTTGTCAATCTTACTTCGGTTGTGTACAATGTATT
TCTGGTCCATTAGGTATGTACCGTAACTCTTTATTACACGAATTTGTAGAAGATTGGTACTCTCAAGAATTTATGGGTAACCAGTGCAGCTTCGGTGATGATCGT
CACTTAACAAACCGTGTATTATCTTTAGGTTACGCTACAAAATACACAGCTCGTTCTAAATGTTTAACAGAAACACCAATTGAATACTTACGTTGGTTAAACCAA
CAAACACGTTGGTCTAAATCTTACTTCCGTGAATGGTTATACAACGCTATGTGGTTCCACAAACACCACTTATGGATGACATACGAAGCGGTAATAACAGGCTTC
TTCCCATTCTTTTTAATTGCTACAGTAATTCAATTATTCTACCGTGGTAAAATTTGGAACATTTTATTATTCTTATTAACAGTACAATTAGTAGGTTTAATAAAA
TCTTCTTTCGCTTCTTGTTTACGTGGTAACATTGTAATGGTATTCATGTCTTTATACTCTGTATTATACATGTCTTCTTTATTACCAGCTAAAATGTTCGCTATT
GCTACAATTAACAAAGCTGGTTGGGGTACATCTGGTCGTAAAACAATTGTAGTAAACTTCATTGGTTTAATTCCAGTATCTGTATGGTTCACAATTTTATTAGGT
GGTGTAATTTTCACAATTTACAAAGAATCTAAAAAACCATTCTCTGAATCTAAACAAACAGTATTAATTGTAGGTACATTATTATACGCTTGTTACTGGGTAATG
TTATTAACATTATACGTAGTATTAATAAACAAATGTGGTCGTCGTAAAAAAGGTCAACAATACGATATGGTATTAGATGTATAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Protein expression successful
 Results
Results
- No catalytic activity probably due to misfolding and intracellular accumulation
Pathway engineering for optimisation of hyaluronic acid production
Number: BBa_K1469002
Description:
UDP-glucose 6-dehydrogenase of Bacillus megaterium. The enzyme catalyses the NAD dependent oxidation of UDP-glucose to UDP-glucuronic acid, a key precursor molecule for hyaluronic acid production.
Sequence:
ATGACTAATATCACAGTAGCGGGTACTGGTTACGTAGGCTTAGTTACAGGTGTTTGTTTAGCTGAAATAGGGCATGAGGTAACATGTGTAGATAA
TATTCAGGATAAAATTGATATTTTAAATGAAGGAATATCTCCTATCTTTGAACCTGGGCTTGATGAATTAATTATTAAAAACTCTGATGCTGGTA
AATTATTTTTCACAACTGATTATCAAAATACTTATGCAAAATCTGATGTTATTTTTATAGGTGTAGGAACACCTGAAAATGAAGATGGGTCTGCT
AATTTAAATTATGTTTATACAGTTGCACAACAAATAGCTAAATTTATTGAGCGAGATTGTCTTATTGTAGTAAAATCTACTGTTCCTATTGGAAC
AAATGAGAAAATAGAAAAATTAATTAAACAACATTTAAAAAATGATGTACAAGTTGAGGTAGCATCCAACCCTGAGTTTTTAGCACAAGGAACAG
CAGTGCGTGACACACTTTATGCATCACGTATTGTTATAGGAGTAGAAAGTGAAAGAGCAAGACAAATATTAACTGATATTTATCAACCTTTTCAT
CAACCTATCTTAACAATGAATCGTCGTAGTGCTGAGATGGTAAAGTATGCTTCAAATGATTTCTTAGCATTAAAAATATCATTTGTTAATGATAT
TGCTAACCTGTGTGAAGTAGTAGGGGCTAACATTGAAGATGTTACAAATGGTATGAGCTATGATCAACGTATTGGAGACAAGTTTTTAAATCCTG
GCATCGGTTACGGTGGTTCATGCTTCCCTAAAGATACTAAAGCGTTACATTGGTTGTCTGAAGAGGAAGGCTATGTATTACGTACCATAAAAGCA
GCTATAGAAGTAAATGAAAAACAAAAGTTTAAACTGATTAAAAAAGCAAGACAAGACTTTGCAACTTTCAAGGGTTTAAAAGTTGCTGTTTTAGG
TCTTACATTCAAGCCTGGTACCGATGACTTACGGGAGGCACCATCAATTCCCAATATTCGTTTATTATTAAATGAAGGTGCTGAAGTTCATGCAT
ATGATCCTGTAGGTGTAAACAATTTTAAGAAAATATATCCAACACAAGTGTATTATGAAGAAACAATTGAAAAAGCATTAAAAAATGCTGACATT
TGTTTTATATTTACAGAGTGGGAAGAAATAAAAAATATGAATTTAAACTTGTTTAACAAACAAATGAAATCTCCTTTTGTCTATGATGGACGTAA
TTGTTACAATCTCGATGAAGTTAAAAGAAGTGGTATCAATTATCAATCTATTGGTCGTCCTAATATAAATGGATACAAAGCTAAAATTAAAGCTA
ATGTCTAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Protein expression successful
 SDS PAGE
SDS PAGE
- characterisation of catalytic activity not possible because no HA production for comparison of HA yields after co expression with hyaluronan synthase 2.
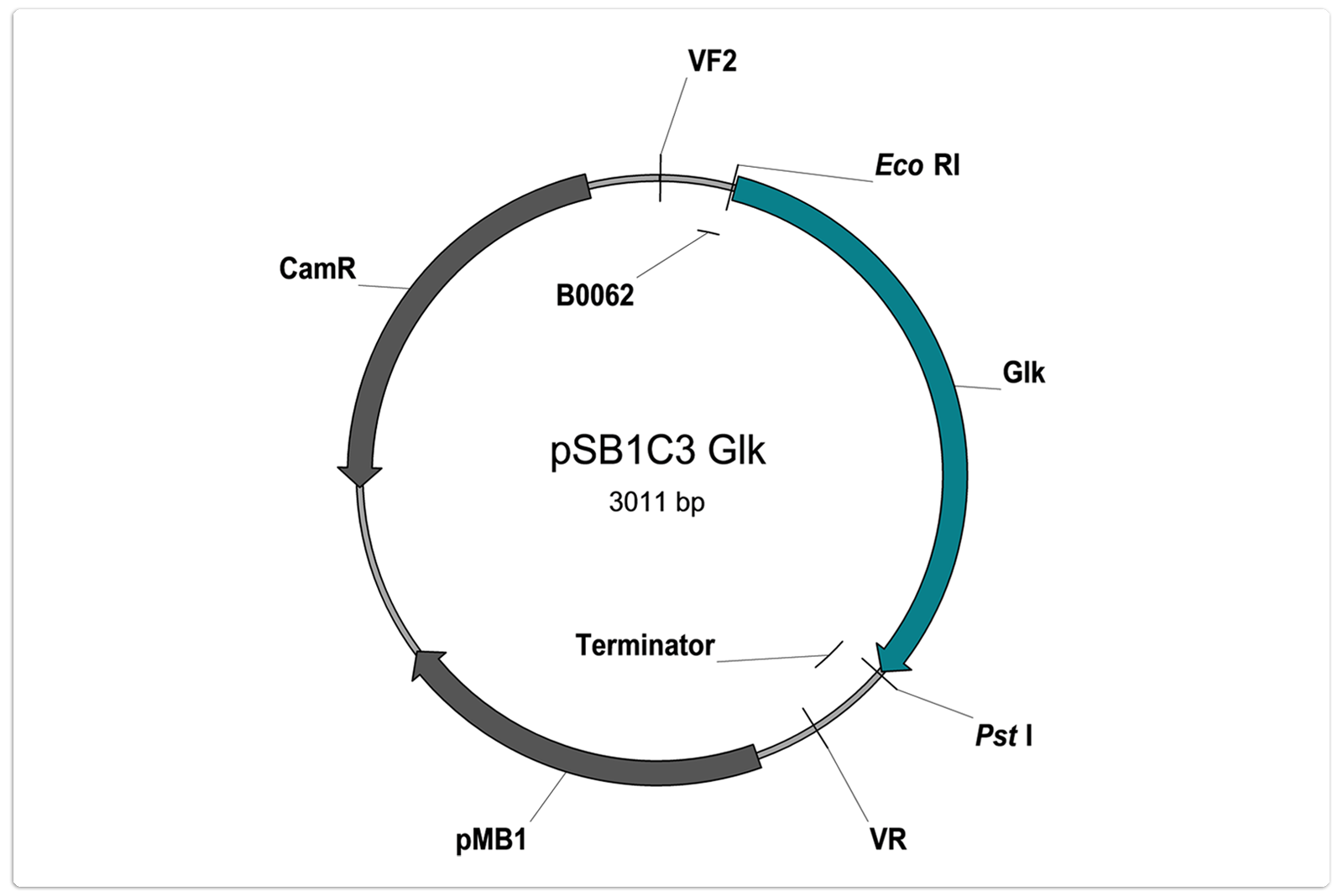
Number: BBa_K1469003
Description:
Glucokinase of Bacillus megaterium. The enzyme catalyses the phosphorylation reaction of glucose to glucose 6-phospahate.
Sequence:
ATGGATGACAAATGGTTAGTTGGAGTTGATTTAGGCGGTACAACAATAAAAATGGCCTTTATTAATCATTATGGAGAAATCATTCACAAGTGGA
AATTAATACGGATGTGAGCGAGCAAGGCCGTAAAATTCCAACGGATATTGCAAAAGCAATTGATAAAAAGTTAAACGATCTTGGAGAAGTAAAA
TCAAGGTTAGTAGGAATTGGCATTGGTGCACCGGGGCCTGTCAACTTTGCAAACGGTTCGATTGAAGTAGCTGTCAATTTAGGTTGGGAAAAAT
TCCCTATAAAAGATATCTTGGAAGTAGAAACTTCTCTTCCTGTTGTAGTAGACAATGATGCAAACATTGCAGCGATTGGAGAAATGTGGAAGGG
TGCTGGAGACGGAGCAAAAGATTTACTTTGCGTTACGCTTGGCACAGGCGTTGGCGGTGGCGTCATTGCAAACGGTGAAATTGTACAAGGCGTA
AATGGAGCCGCTGGTGAGATCGGGCACATTACTTCTATTCCTGAAGGCGGGGCACCGTGTAACTGCGGTAAAACCGGCTGTTTAGAAACCATTG
CTTCAGCAACTGGAATTGTACGTTTAACAATGGAAGAATTAACGGAAACGGACAAACCAAGTGAGCTTCGCACAGTGTTAGAACAAAATGGACA
AGTTACATCTAAAGATGTATTTGATGCAGCTCGTTCAAAAGACGGGTTAGCTATGCATGTTGTAGATAAAGTTGCTTTTCATTTAGGTCTAGCA
CTAGCAAACTCTGCTAATGCATTAAACCCTGAGAAGATCGTTCTAGGCGGCGGTGTGTCTCGTGCAGGCGAGGTATTACTTGCACCGGTAAGAG
ATTATTTCAAACGTTTTGCATTTCCTCGCGTAGCGCAAGGTGCTGAACTAGCAATCGCTACTTTAGGAAACGATGCGGGAATTATTGGAGGAGC
TTGGTTAGTTAAATCTTATTTTGAATAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform.
- As indicated on the vector map: GCGGCCGCTTCTAG of prefix and TACTAGTAGCGGCCG of suffix are deleted
- Use only after amplification with appropriate primers
Number: BBa_K1469004
Description:
Beta-phosphoglucomutase of Bacillus megaterium. The enzyme catalyses the conversion of glucose 6-phospahate to glucose 1-phosphate.
Sequence:
ATGAAAAAAGAAGAGCTACAAGCGATTATTTTTGATTTAGACGGTGTTATTGCAGATACGGTACATTTATATTATAAAGCAAATAAAAAAGT
AGCAGATCAACTTAACGTTTCATTTTCTGAAGAACTCAATCAGCAGCTGCAGGGGATCAGCCGAGTGAAAACTGTAGAGCTCATTGCTGCCC
AAGGCAATGTACATCTGACAATGGAAAAAAAAGAACAGCTTGCTGATGAAAAGAATAAGCATTATCAAGCATTAATAAAGGAAATGACAAAG
GATCATCTACTTCCAGGAATACAATTGTTGTTAGAGGATTGTAAAAAACAAGGTATTAAAATGGCGATTGCTTCTTCTAGTTCAAATGCAAA
TACTGTAGTGGATGCGTTAGGAATACGTCCTTTTTTTGATTGCATTGTGGATGTGAGAAAAATAAAGAAGGGAAAACCAGACCCAGAAATAT
TTTTGACAGCTGCCCATCAATTAAAAGTAACACCCTATGCATGTGTGGCTATAGAAGATGGAGAGGCGGGTGTAAAAGCTATTAATCAAACA
GATATGGTTTCTATCGGCGTTGGTAAGCATCTGAAGGAAGAGGATATGAATTTTCACGTATATTCAACAGCTGATTTGCATTTAAAGACGAT
CAAAGAGGTTTTCGAAAAAACAAGAAAAGGAACAGATTATAACAGCGTGTATACAAAAAATTAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: GCGGCCGCTTCTAG of prefix is deleted
- Use only after amplification with appropriate primers
Number: BBa_K1469005
Description:
UTP--glucose-1-phosphate uridylyltransferase of Bacillus megaterium. The enzyme catalyses the conversion of glucose 1-phospahate to UDP-glucose 1-phosphate.
Sequence:
ATGACGATAAAAAAGGCAGTTATTCCAGTAGGCGGTTTAGGAACTCGATTTTTGCCAGCGACAAAAGCGCAGCCCAAAGAAATGCTGCCAAT
TGTCGATAAGCCTGCTGTACAATATATTGTAGAAGAAGCTGTGAAATCAGGTATTGAAAGTATTATTTTTATTACAGGACGAAACAAAAAAT
CAATTGAAGACCATTTTGATAAATCAACTGAGTTAGAGCAAATGCTTGAAGAAAAACACAAGTTCCAAATGCTAAAAGAAGTACAAACGATC
TCATCAATGGCCAGCATTCACTATATTCGTCAAAAAGAGCCGTTAGGACTTGGTCATGCTATTCTTTGTGCCGAACAGTTTATTGGAGATGA
GCCGTTTGCCGTTCTTCTTGGAGACGATATTATGACTTCTGAAGAACCTGCGCTCAAACAGATAATCCAAGCGTATGAAACGACTAATCAAT
CAGTTATCGGAGTTCAAAAAGTGGATGTTGAAGAGGTATCAAAATACGGAATCATTCAGCCGAAAAATAAATCTGGCAGCCTTCATGAAGTG
AATGACTTGATTGAAAAACCATCGATAGAACAAGCTCCGTCAACTATTGCCGTTATGGGCCGCTATATTTTAAATCCTTCTATTTTTTCTTA
TCTAAAAACGATTGAAAGAGGGGTAGGAAATGAATTACAATTAACAGATGCACTGGGTGTGGTATGTGAAAAAGAGCGATTGTTTGCTCTTG
AACTGGAAGGACAGCGCTTTGATATTGGAGATAAGGTAGGCTATATGAAAGCGATGGTAGAAGTTGCGTTAAAGCGCAGAGATTTACGTCAA
ACGTTTTTATCGTATTTGGAGGGAATTGTAGAAAAAGAACGAGCAAACAACTTCAGCTAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: GCGGCCGCTTCTAG of prefix and TACTAGTAGCGGCCG of suffix are deleted
- Use only after amplification with appropriate primers
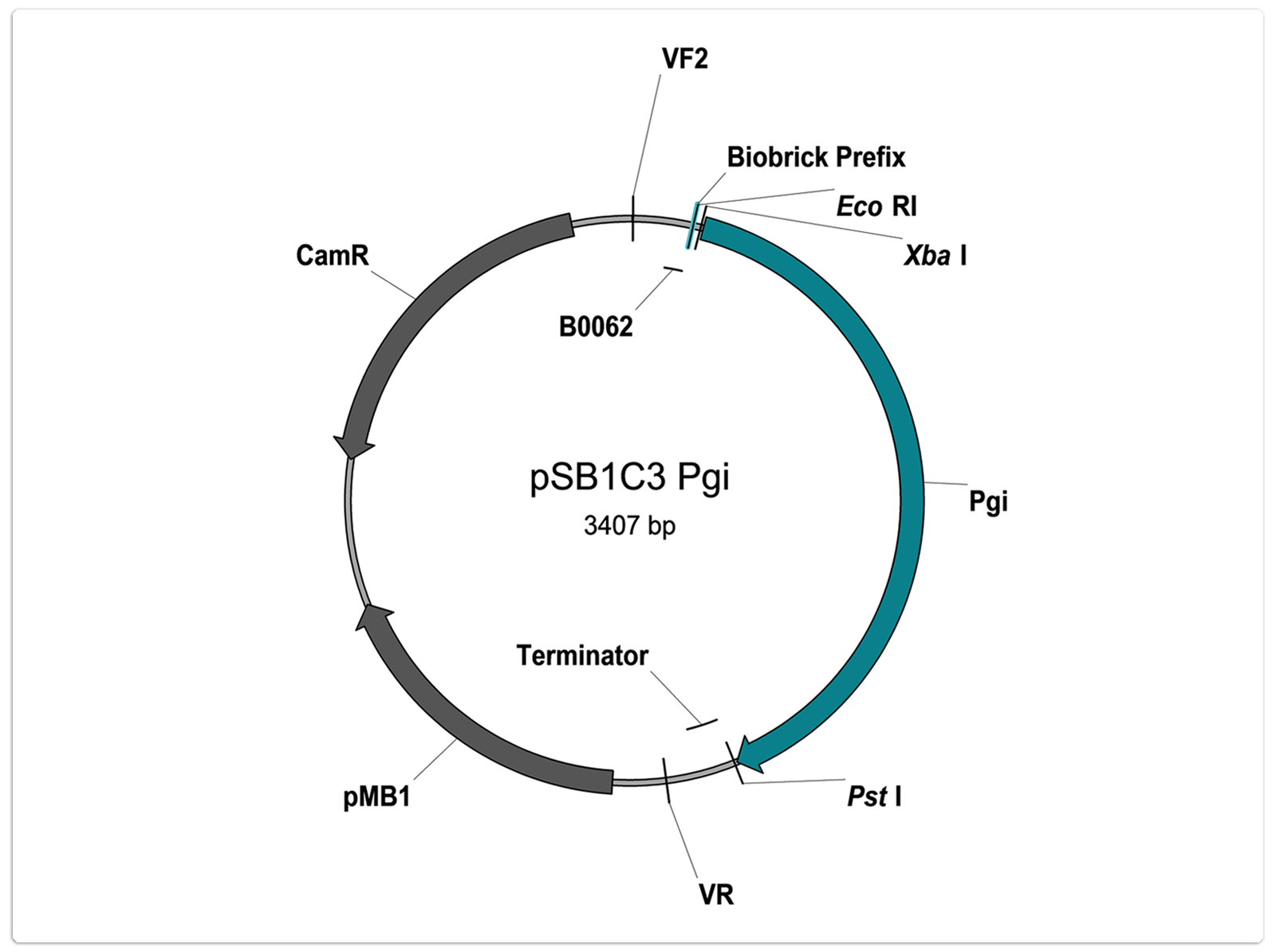
Number: BBa_K1469006
Description:
Glucose-6-phosphate isomerase of Bacillus megaterium. The enzyme catalyses the conversion of glucose 6-phospahate to fructose 6-phosphate.
Sequence:
ATGACACATGTTCGTTTTGATTATTCAAACGCGTTATCATTTTTTGGTGAACATGAACTTACATACTTACGTGATGCCGTGAAAGTAGCTC
ACCATTCAATTCATGAGAAAACAGGTGCTGGAAACGACTTTTTAGGATGGGTCGACCTTCCTACGGAGTATGACAAAGAAGAATTTGCACG
CATTCAAAAAAGTGCAGAAAAAATTAAATCTGATTCAGATGTATTAATTGTTATCGGTATTGGTGGATCTTACTTAGGTGCTCGTGCTGCT
ATTGAAATGCTTCAGCATTCATTCTATAACACATTATCAAAAGAACAGCGTAAAACTCCGCAAGTTATTTTTGCTGGTAACAACATCAGCT
CAACATACATGAAAGACTTAATGGATGTTTTACAAGATAAAGATTTCTCTATTAATGTTATTTCAAAATCTGGTACAACAACAGAACCGGC
GATTGCTTTCCGTATCTTCCGTAAATTATTAGAAGAGAAATACGGCAAAGAAGAAGCACGCAAGCGTATTTATGCAACAACAGATAAAGCG
CGCGGTGCATTAAAAACATTAGCGGACGAAGAAGGCTATGAAACATTTGTTATTCCAGATGATGTAGGCGGCCGTTATTCAGTATTAACAG
CGGTTGGCTTATTACCAATTGCCGCTGCGGGTCTTGATATCGAACAAATGATGAAAGGGGCAGCTGATGCTCAAGAAGCATTCTCATCATC
AGAGCTAGAAGAGAACCCAGCTTATCAATATGCTGTAGCTCGTAATGTGCTTTATAATAAAGGAAAAACAATTGAAATGCTGATTAACTAT
GAGCCAGCTCTTCAATACTTTGCTGAGTGGTGGAAACAGTTATTTGGCGAAAGTGAAGGAAAAGACCAAAAAGGTATTTACCCTTCATCTG
CTAACTTCTCAACGGATCTTCATTCTTTAGGTCAATATGTACAAGAAGGTCGCCGTGATATTTTTGAAACAATTGTACAAGTTCAAGATTC
TCGTCATGAGCTAACAATTGAAGAAGATGCAGCGGACTTAGATGGATTAAATTACTTAGCTGGTGAAACAGTTGACTTTGTAAATAAAAAA
GCGTTCCAAGGTACAATGCTTGCTCACACAGATGGAGGCGTTCCAAACTTAGTTGTGAACATTCCTGCGCTAGATGAGTATACGTTCGGAT
ATGTTGTATATTTCTTCGAAAAAGCATGTGCAATCAGCGGCTATTTATTAGGCGTAAACCCATTCGATCAGCCGGGAGTAGAAGCATACAA
AGTAAATATGTTTGCTCTTCTTGGAAAACCTGGTTTTGAAGAGAAAAAAGCAGAATTAGAAAAGCGTTTGAAATAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: TACTAGTAGCGGCCG of suffix is deleted
- Use only after amplification with appropriate primers
Number: BBa_K1469007
Description:
Glucosamine--fructose-6-phosphate aminotransferase (isomerising) of Bacillus megaterium. The enzyme catalyses the conversion of fructose 6-phosphate to glucosamine 6-phosphate.
Sequence:
ATGTGCGGAATTGTAGGATATATTGGAACAGAAGATTCGAAAGAAATTTTATTACGTGGTCTTGAAAAGTTAGAATATCGTGGCTATGACT
CAGCGGGTATTGCAGTTGTTAACAATGAAGGCGTACACGTATTCAAAGAAAAAGGCCGTATTGCAGAATTGCGTCAGGCAGTAGATAACAG
TGTTGTAGCACCAGCAGGTATCGGTCATACGCGTTGGGCAACTCATGGTGTACCAAGCCAAGAAAATGCTCACCCGCATCAAAGTACATCT
GGACGCTTCACACTTGTGCATAACGGTGTAATTGAAAACTATGCTATTTTGAAACGTGAATATTTACAAGATGTAACGTTCAAAAGCGAAA
CGGATACAGAAGTAATCGTTCAGTTAATTCAAAAAATCGCAGCAGAAGGCTTAGATGTAGAAGAAGCGTTCCGTAAGACAGTAAGCTTGCT
TCATGGTTCTTATGCACTAGCTCTTGTTGATAACAAAGATAAAAATACAATCTACGTAGCTAAAAACAAAAGCCCGCTTCTTGTAGGTGTT
GGTGAAGGATTTAACGTTGTAGCGAGTGATGCTATGGCAATGCTTCAAGTAACAGATCAGTACGTAGAATTAATGGATAAAGAAACAGTAA
TTGTTACAAAAGAAGGCGTAACAATTAAAACGCTTGATGGTGAAGTGGTAGAAAGAGCACCTTATACAGCAGAGCTTGATGCAAGTGATAT
TGAAAAAGGTACGTACCCTCACTATATGTTAAAAGAAATTGACGAGCAGCCTTTAGTTGTTCGTAAAATCATTCAAGAGTATCAAAATGAG
AACAATGAACTGCACATTGATGCAGACATTTTAGGAGCAATGGATGAAGCAGACCGCATCTATATCGTAGCATGTGGAACAAGCTATCATG
CAGGCCTAGTAGGTAAACAATTCATTGAAACGTGGGCGAAGGTTCCAGTAGAAGTGCACGTAGCAAGTGAATTTTCTTATAACATGCCGCT
TTTATCTGAAAAGCCATTGTTTATCTTTATTTCTCAAAGTGGAGAAACAGCTGACAGCCGTGCAGTATTAGTTCAAATTAAAGAACTAGGT
TACAAAGCGTTAACAATCACAAACGTTCCGGGATCAACATTATCTCGTGAATCTGATTATACATTATTACTTCATGCAGGTCCGGAAATTG
CTGTAGCATCAACAAAAGCTTATACAGCTCAATTAGCAGTGCTATCAATTCTAGCGGCTGTAACAGCAAAAACACGCGGCATTGAATTAGA
ATTTGATTTAGTTCAAGAGTTAGCAATCGTAGCAAACACAATGGAAGTACTTTGCGACGACAAAGAAGAAATGGAAAGCATTGCTCTTCAA
TTCTTAGCAACAACTCGCAACGCATTCTTTATTGGACGTTCTGTCGATTATTATGTAGGTTTAGAAGGTGCGTTAAAACTAAAAGAAATCT
CTTACATCCAAGCAGAAGGATTTGCTGGAGGAGAGCTTAAGCACGGAACAATCGCTTTAATTGAAAACAATACGCCAATTATTGCATTAGC
AACACAAGAGCATGTAAACTTAAGCATCCGTGGTAACGTAAAAGAAGTAGTAGCACGTGGAGCTAATCCTTGTATCATTTCAATGAAAGGA
TTAGAAACAGAAGAAGATCGCTACGTGATTCCGAAAGTTCACGAAACACTTTCACCACTAGCAGCGGTTATTCCTTTACAATTAATCTCTT
ACTACGCTGCATTGCACCGCGATTGCGATGTAGATAAACCACGTAACTTAGCTAAATCAGTTACTGTAGAATAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: TACTAGTAGCGGCCG of suffix is deleted
- Use only after amplification with appropriate primers
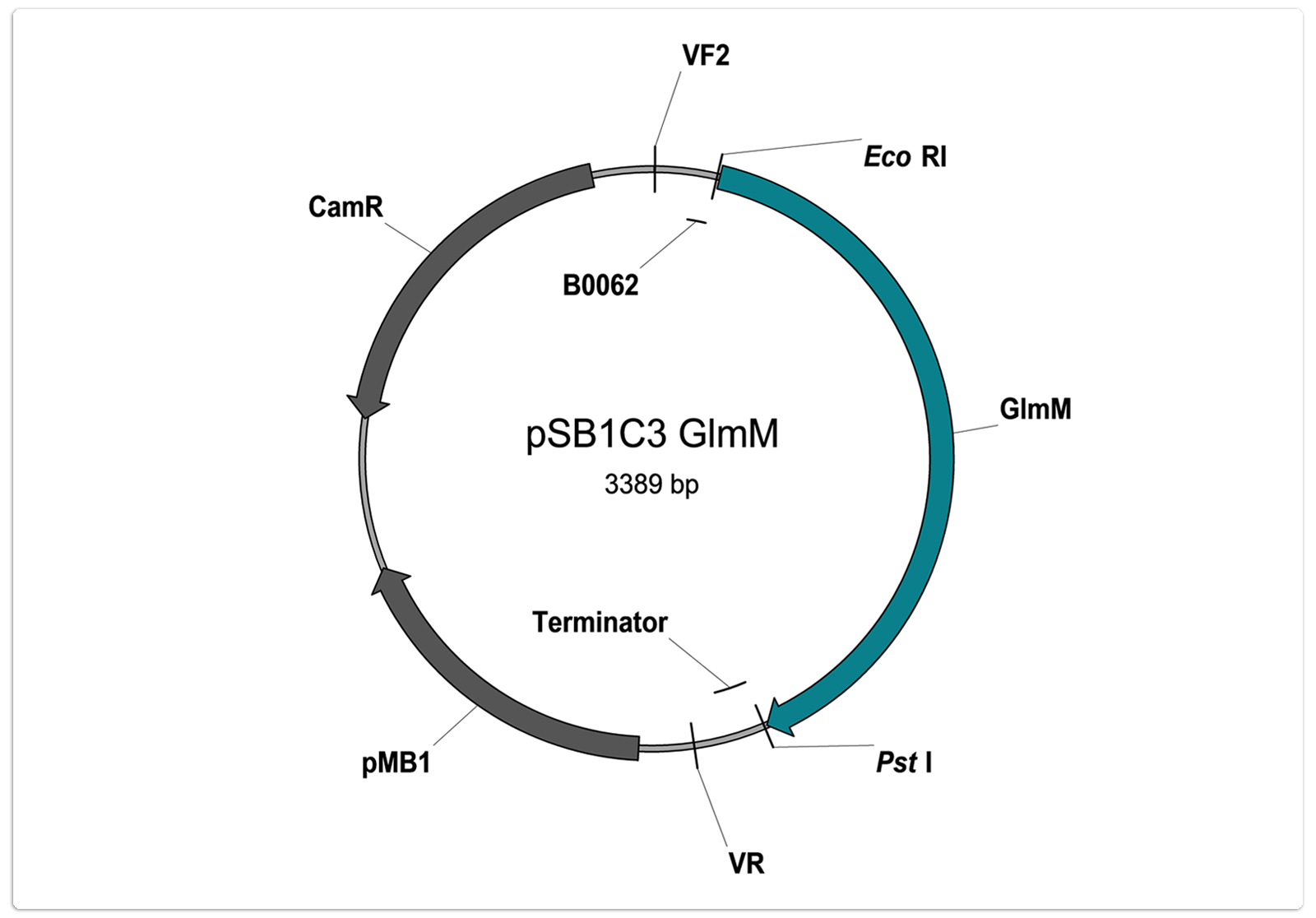
Number: BBa_K1469008
Description:
Phosphoglucosamine mutase of Bacillus megaterium. The enzyme catalyses the conversion of glucosamine 6-phosphate to glucosamine 1-phosphate.
Sequence:
ATGGGTAAGTATTTTGGAACAGACGGAGTACGAGGAGTTGCCAATAGTGAATTAACTCCTGAGCTTGCTTTTAAAATTGGACGCTTAGGTG
GTTACGTTCTAACAAAAGATTCAGAGCGACCAAAAGTATTAATCGGACGCGATACACGCGTTTCTGGTCATATGTTAGAAGGAGCGCTTGT
TGCAGGATTGCTTTCAATTGGAGCAGAAGTTATGCGACTAGGTGTTATTTCAACTCCAGGGGTAGCGTATTTAACAAAAGCACTTGGCGCA
CAAGCAGGAGTCATGATTTCTGCTTCGCATAACCCAGTTCAAGATAACGGAATTAAATTCTTTGGTCCAGATGGATTTAAACTTTCAGATG
CCCAAGAGAATGAAATTGAAGCGTTAATGGATAGTGACACAGATCAATTACCAAGACCTGTTGGGGGAGACTTAGGTCAGGTAAATGACTA
TTTCGAAGGTGGACAAAAATATCTTCAATACTTAAAACAAACGGTTGATGAAGACTTCTCAGGTATTCATGTGGCATTAGACTGTGCGCAC
GGGGCAACGTCATCGTTAGCAGCACATTTATTTGCAGATCTCGATGCAGATATCTCTACAATGGGGGCATCTCCAAACGGATTGAACATCA
ATGACGGTGTAGGATCTACACATCCGGAAGCATTAAGTGCCTTCTTAAAAGAAAAAGGAGCGGATGTTGGATTAGCATTTGATGGAGACGG
AGATCGTTTAATCGCAATCGACGAAAAAGGCGAGATTGTAGACGGTGATCAAATTATGTTCATTTGTGCAAAATACTTATATGCACAAGGT
CGTTTAAAGAAAAACACGCTTGTTTCTACCGTTATGAGTAACTTAGGCTTCTATAAAGCACTAGAAGCTAGCGGGATTGAAAGTGCACAAA
CTGGTGTAGGAGACCGTTATGTAGTAGAAGAAATGAAAGCTAATCAATATAACCTAGGTGGAGAACAGTCAGGTCATATTATTTTCTTAGA
TTACAACACAACAGGTGACGGTATGCTTTCAGCTATTCAGCTTGTAAACATTATGCAAGCAACGAAAAAGCCTTTATCAGAATTAGCAGCT
GAAATGAAGAAATATCCTCAGCTTCTTGTAAATGTAAAAGTAACAGATAAGCATCATGTTACAGATAACGAGAAGGTAAAAGTGGTTATTG
AAGAAGTGGAAAAAGAAATGAACGGTAATGGCCGAGTACTTGTACGCCCTTCAGGAACAGAGCCGTTAGTACGTGTGATGGTTGAAGCACA
AACACAAGAAGAATGTGAAACTTATGTAACGCGTATCGTAGAAGTAGTAAAAGAAGAAATGGGCTTAGAGTAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: GCGGCCGCTTCTAG of prefix and TACTAGTAGCGGCCG of suffix are deleted
- Use only after amplification with appropriate primers
Number: BBa_K1469009
Description:
UDP-N-acetylglucosamine pyrophosphorylase of Bacillus megaterium. The enzyme is bifunctional and catalyses the conversion of glucosamine 1-phosphate to N-acetyl glucosamine 1-phosphate as well as the conversion of N-acetyl glucosamine 1-phosphate to UDP-N-acetyl glucosamine, a key precursor molecule for hyaluronic acid production.
Sequence:
ATGTCAAAAAGATATGCAGTCATATTGGCAGCTGGTCAGGGTACACGTATGAAGTCATCTTTATATAAAGTGTTACACCCTGTGTGTGGCAA
ACCGATGGTGCAACACGTAATTGATCAGTTATCACGTTTGGATTTAACAAGTTTAATTACAGTTGTAGGTCACGGTGCCGAAAAAGTGAAAT
CACATGTTGGAGATAAAAGCTTATTTGCTTTACAAGAAGAGCAGTTAGGAACAGCTCATGCAGTAATGCAAGCAGAATCTTTACTTGCACAT
GAAAAAGGTACGACAATTGTTGTATGTGGAGATACTCCGCTTATTACAGCTGAAACAATTGAATCTTTATTAAAGCATCATGAAGCAACAAA
TGCAATGGCAACTATTTTAACAGCTCATGCAGAAGATCCAACAGGATATGGACGCATCATTCGAAATGAACAAGGGTCTGTTGAAAAGATTG
TGGAGCATAAAGATGCCACAGAACAAGAGCGTCTTGTACAGGAAATTAATACTGGTACGTATTGTTTTGATAATGAGAAATTATTTGAGACA
CTTTCAAAAGTTTCAAATGATAATGTACAGGGAGAGTATTATTTACCTGATGTTATCGAAATTCTAAAAAATGAAGGTCAAATTATTTCTGC
ATATCAAACATCCGATTTTGCTGAAACATTGGGCGTTAATGATCGATTTGCTCTGTCTCAAGCAGAAGAAACAATGAAAAAGCGCATCAACA
AGCAGCATATGTTAAATGGAGTGACGATTGTGGATCCTTCTAACACGTACATTTCAGCAGATGCTGTCATTGGCAGAGATACGGTTATTTAT
CCTGGTACGGTTATTCAAGGAACAGTTGTAATTGGAGAGAATTGTGAAGTAGGACCTAACAGTGAAATTAAAGACTGCAAAATAGGTAATAA
TACTTCTATTCGCCATTCTGTTGCTCACGATAGTGAAATTGGTCACGAAGTCACAATTGGCCCATTTGCACATATTCGTCCTCAATCATTAA
TTGGAGATGAAGTGCGCGTAGGAAACTTTGTAGAAATTAAAAAAGCATCTTTTGGCAAAGGTAGCAAAGCTTCTCATTTAAGTTATATTGGA
GACGCAGAGGTCGGAAAAGGTGTAAACTTAGGGTGCGGATCTATTACAGTTAACTACGATGGGAAAAATAAATTTTTAACAAAGATTGAAGA
TGGGGCATTCGTAGGATGTAACTCAAACTTAATTGCACCTGTAACAATTGGTGAAGGCGCCTATGTTGCAGCTGGTTCAACCGTAACGGATG
ATGTGCCAGGAAAAGCATTATCAATTGCTCGTGCGAAACAAGTAAATAAAGAAAACTATGCTGAAAAGCTTGATATTAATAAAAAATCCTAA
Characterisation status:
- Part is user sequenced: no illegal restriction sites, no mutations
- Part is not RFC10 conform
- As indicated on the vector map: GCGGCCGCTTCTAG of prefix and TACTAGTAGCGGCCG of suffix are deleted
- Use only after amplification with appropriate primers
 "
"









Impressum/Copyright