Team:LMU-Munich/Project/B subtilis
From 2014.igem.org
Bacillus subtilis
We aim to introduce quorum sensing modules from the two Gram-positive pathogens Staphylococcus aureus and Streptococcus pneumoniae into an appropriate chassis that will sense and eventually kill these pathogens specifically.
As Bacillus subtilis is a Gram-positive rod shaped bacterium and many of us had good experiences working with this microorganism, we took B. subtilis as our chassis of choice. Furthermore, the guanine-cytosine contents of B. subtilis (43.5 %), S. aureus (32.8 %) and S. pneumoniae (39.7 %) are comparable, which is of great advantage for heterologous gene expression. In some cases optimizing the codon usage of the gene of interest derived from another organism is very helpfull: We developed a tool to perform codon adaptation on your own, without the need to pick each matching codon by hand .
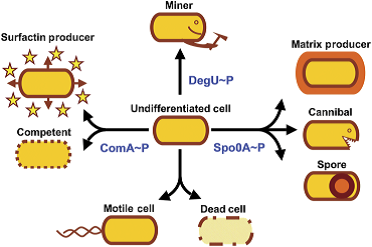
B. subtilis is a soil bacterium that has to face changing environmental stimuli like temperature, pH or scarcity of nutrients. In order to cope with this, B. subtilis is able to differentiate into different cell types. [http://www.ncbi.nlm.nih.gov/pubmed/20030732] During starvation, B. subtilis is able to kill his siblings to gain new nutrition by introducing specific toxins to the extraplasmic space. If the conditions get even worse, the soil bacterium is able to form long living, stable endospores. However, if nutrients become available again, they are able to germinate and re-enter the vegetative cycle.
Furthermore, our model organism is a nonpathogenic and nontoxicogenic strain and was declared as GRAS (Generally Regarded As Safe) by the Food and Drug Administration [http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/MicroorganismsMicrobialDerivedIngredients/default.htm (FDA)]. In our lab we use the tryptophan-requiring, auxotrophic B. subtilis strain 168, whose genome was completely sequenced in the year 1997 [http://www.ncbi.nlm.nih.gov/pubmed/9384377] [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2580678/].
In addition, the iGEM Team LMU-Munich 2012 has constructed the Bacillus BioBrickBox (B4), which contains several well evaluated integrative and replicative vectors for B. subtilis , thus providing a powerful toolbox for the engineering of B. subtilis. One extraordinary aspect of B. subtilis is its ability to perform natural transformation in its post-exponential phase of cell growth. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC372826/]. This feature can be used in the lab very reliably. For example, integrative B4 vectors are constructed so that the DNA fragment of interest is flanked by homologous regions to the B. subtilis genome. Thus, via homologous recombination at this locus, B. subtilis will integrate the DNA-fragment and replicate it with his own chromosome.
Due to all of these helpful features, B. subtilis is our chassis organism of choice! Check out our results, obtained with this organism!
Sources
[1], [2] D. Lopez, R. Kolter: Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. (2010)134-49
[3] Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF et all: The complete genome sequence of the gram-positive bacterium Bacillus subtilis (1997). Nature 249-56
[4] Daniel R. Zeigler, Zoltán Prágai, Sabrina Rodriguez, Bastien Chevreux, Andrea Muffler, Thomas Albert, Renyuan Bai, Markus Wyss, and John B. Perkins: The Origins of 168, W23, and Other Bacillus subtilis Legacy Strains. J Bacteriol. 190(21): 6983–6995.
[5] D Dubnau: Genetic competence in Bacillus subtilis. (1991) Microbiol Rev.; 55(3): 395–424.
 "
"