Team:EPF Lausanne/Overview
From 2014.igem.org
Arthurgiroux (Talk | contribs) |
|||
| (182 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{CSS/EPFL_head}} | {{CSS/EPFL_head}} | ||
<html> | <html> | ||
| - | + | <style> | |
| - | + | #contentSub, #footer-box, #catlinks, #search-controls, #p-logo, .printfooter, .firstHeading,.visualClear {display: none;} /*-- hides default wiki settings --*/ | |
| - | + | </style> | |
| - | + | ||
| - | + | <!-- here ends the section that changes the default wiki template to a white full width background --> | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- MENU --> | ||
<nav class="navbar navbar-default navbar_alt" role="navigation"> | <nav class="navbar navbar-default navbar_alt" role="navigation"> | ||
<div class="container-fluid"> | <div class="container-fluid"> | ||
| Line 26: | Line 31: | ||
<li><a href="https://2014.igem.org/Team:EPF_Lausanne">Home</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne">Home</a></li> | ||
<li class="dropdown"> | <li class="dropdown"> | ||
| - | <a href="#" class="dropdown-toggle" data-toggle="dropdown">Project <span class="caret"></span></a> | + | <a href="#" class="dropdown-toggle active" data-toggle="dropdown">Project <span class="caret"></span></a> |
<ul class="dropdown-menu" role="menu"> | <ul class="dropdown-menu" role="menu"> | ||
| - | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Overview">Overview</a></li> | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Overview" class="active">Overview</a></li> |
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria">Stress Responsive Bacteria</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria">Stress Responsive Bacteria</a></li> | ||
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/Yeast">Osmo Responsive Yeast</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Yeast">Osmo Responsive Yeast</a></li> | ||
| Line 38: | Line 43: | ||
</li> | </li> | ||
<li class="dropdown"> | <li class="dropdown"> | ||
| - | <a href="#" class="dropdown-toggle | + | <a href="#" class="dropdown-toggle" data-toggle="dropdown">Achievements <span class="caret"></span></a> |
<ul class="dropdown-menu" role="menu"> | <ul class="dropdown-menu" role="menu"> | ||
| - | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Results | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Results">Results</a></li> |
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/Data">Data</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Data">Data</a></li> | ||
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/Judging">Judging</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Judging">Judging</a></li> | ||
| Line 47: | Line 52: | ||
</li> | </li> | ||
<li class="dropdown"> | <li class="dropdown"> | ||
| - | <a href="#" class="dropdown-toggle" data-toggle="dropdown">Policy & | + | <a href="#" class="dropdown-toggle" data-toggle="dropdown">Policy & Practices <span class="caret"></span></a> |
<ul class="dropdown-menu" role="menu"> | <ul class="dropdown-menu" role="menu"> | ||
| - | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/HumanPractice">Human | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/HumanPractice">Human Practices</a></li> |
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/Safety">Bio Safety</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Safety">Bio Safety</a></li> | ||
<li><a href="https://2014.igem.org/Team:EPF_Lausanne/PolicyPractice">Metafluidics</a></li> | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/PolicyPractice">Metafluidics</a></li> | ||
| Line 78: | Line 83: | ||
</div><!-- /.container-fluid --> | </div><!-- /.container-fluid --> | ||
</nav> | </nav> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <!-- END MENU --> | |
| - | + | ||
| - | + | <!-- ABSTRACT --> | |
| - | + | ||
| - | + | ||
| - | + | <div class="container"> | |
| - | + | <div class="box" id="boxbread"> | |
| - | + | ||
| - | + | <ol class="breadcrumb breadcrumb-arrow"> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne"><i class="glyphicon glyphicon-home"></i> Home</a></li> | |
| - | + | <li class="dropdown"><a href="#" class="dropdown-toggle" data-toggle="dropdown"><i class="glyphicon glyphicon-cog"></i> Project <b class="caret"></b></a> | |
| - | + | <ul class="dropdown-menu"> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria">Stress Responsive Bacteria</a></li> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Yeast">Osmo Responsive Yeast</a></li> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Microfluidics">Microfluidics</a></li> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Hardware">Hardware</a></li> | |
| - | + | <li><a href="https://2014.igem.org/Team:EPF_Lausanne/Applications">Applications</a></li> | |
| - | + | </ul> | |
| - | + | </li> | |
| - | + | <li class="active"><span><i class="glyphicon glyphicon-picture"></i> Overview</span></li> | |
| - | + | </ol> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | </ | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| + | |||
| + | <div class="row"> | ||
| + | |||
| + | <div class="col col-md-9"> | ||
| + | |||
| + | |||
| + | <div class="whitebg box"> | ||
| + | <!-- PROJECT --> | ||
| + | |||
| + | <div class="align-justify"> | ||
| + | |||
| + | <h1 class="cntr">Overview</h1> | ||
| + | |||
| + | <h2 class="section-heading" id="title_intro">Introduction</h2> | ||
| + | <p class="lead"> | ||
| + | |||
| + | <!--Our team has been working on showing that biologically engineered organisms can detect and process signals quickly and efficiently.--> | ||
| + | |||
| + | Biological responses can be quick and precise! This is the message that our team wants to convey. With this in mind, our team brought forward a novel idea: | ||
| + | <br /> | ||
| + | |||
| + | <p style="border-style:solid; border-color:#120A2A;padding: 25px; background-color:#A5D2DA"> | ||
| + | Combining Protein Complementation techniques with biosensors and microfluidics allows fast spatiotemporal analysis of bacterial/yeast responses to stimuli. | ||
| + | </p> | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | Biological responses can be quick! This is the message that the 2014 EPFL iGEM team want to convey. | ||
| + | |||
| + | <br /><br /><i>The BioPad project is intended to be a solid proof-of-concept of the combination of biosensors with protein complementation techniques to achieve fast spatiotemporal analysis of bacterial or yeast response to mechanical stimuli.</i> | ||
| + | </p> | ||
| + | --> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p class="lead"> | ||
| + | Our team explored this hypothesis by engineering two stress related pathways in <i>E. coli</i> and <i>S. cerevisiae</i> with in mind the development of a BioPad: a biological touchscreen consisting of a microfluidic chip, touch-responsive bacteria/yeast, and a signal detector. Learn more about <a href="#howitworks">how the BioPad works !</a> </p> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- | ||
| + | <p class="lead"> | ||
| + | The pathway engineered in <i>E. coli</i>, the Cpx Pathway, is a two-component (CpxA and CpxR) regulatory system responsive to envelope stress. It controls the expression of "survival" genes whose products act in the periplasm to maintain membrane integrity. CpxA intermembrane kinase activates and autophosphorylates when sensing misfolded protein (due to stress for example). CpxA phosphorylates CpxR which homodimerizes, acting as a transcription factor. A full description of the pathway is available <a href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria">Envelope Stress Responsive Bacteria page !</a></p> | ||
| + | |||
| + | --> | ||
| + | |||
| + | <p class="lead"> | ||
| + | The pathway engineered in <i>E. coli</i>, the Cpx Pathway, is a two-component regulatory system responsive to envelope stress. A full description of the pathway is available <a target="_blank" href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria#cpx">here.</a> | ||
| + | <br/> | ||
| + | In <i>S. cerevisiae</i> we modified the HOG Pathway - a MAPKK pathway responsive to osmotic stress. For more information concerning the HOG Pathway click <a target="_blank" href="https://2014.igem.org/Team:EPF_Lausanne/Yeast">here.</a> | ||
| + | </p> | ||
| + | |||
| + | <br/> | ||
| + | |||
| + | <div class="cntr"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/f/fd/CpxR-HOG.jpg" data-lightbox="cpxr" data-title="CpxR-HOG"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/f/fd/CpxR-HOG.jpg" alt="touch response" class="img-responsive img-border" /></a> | ||
| + | </div> | ||
| + | |||
| + | <br /><br /><br/> | ||
| + | |||
| + | <div class="pull-left img-left"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/d/d8/EPFLmicrofluidics.JPG" data-lightbox="image-1" data-title="EPFL microfluidic chips"><img src="https://static.igem.org/mediawiki/2014/d/d8/EPFLmicrofluidics.JPG" width="250" class="img-border"></a> | ||
| + | <figcaption class="cntr">EPFL microfluidic chips</figcaption> | ||
| + | </div> | ||
| + | <p class="lead"> | ||
| + | |||
| + | Our project also includes an extensive <b>microfluidics section</b>. Our self designed chips helped us improve precision, safety and quantification methods used throughout the project. To learn more about the microfluidic components of our project check out <a target="_blank" href="https://2014.igem.org/Team:EPF_Lausanne/Microfluidics">this link.</a></p> | ||
| + | |||
| + | <div class="pull-right img-right"> | ||
| + | <img src="http://www.raspberrypi.org/wp-content/uploads/2011/07/RaspiModelB.png" alt="first" width="200" class="img-border"> | ||
| + | <figcaption class="cntr">Raspberry Pi</figcaption> | ||
| + | </div> | ||
| + | <p class="lead"> | ||
| + | <br /> | ||
| + | Last but not least, we designed a <b>novel signal detector</b>! To make signal detection more practical we developed an automatised cheap tracking system made of a mini-computer (Raspberry Pi) and a mini-HD camera. More details concerning the BioPad detector can be found <a target="_blank" href="https://2014.igem.org/Team:EPF_Lausanne/Hardware">here.</a> | ||
| + | <br /></p> | ||
| + | |||
| + | <!-- | ||
| + | <h2 id="CpxPathway">The Cpx Pathway in <i> E.Coli </i></h2> | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | <!-- CpxA-CpxR PATHWAY DESCRIPTION --> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | <div class="cntr"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/6/62/Cpx_pathway_2_far_2.jpg" alt="Cpx_pathway_description_diagram" class="img-responsive"> | ||
| + | </div> | ||
| + | --> | ||
| + | |||
| + | <!-- | ||
| + | <p class="lead"> | ||
| + | The natural function of the Cpx two component regulatory system in bacteria is to control the expression of ‘survival’ genes whose products act in the periplasm to maintain membrane integrity. This ensures continued bacterial growth even in environments with harmful extracytoplasmic stresses. The Cpx two component regulatory system belongs to the class I histidine kinases and includes three main proteins: CpxA, CpxR, and CpxP. For more detailed information about the Cpx pathway check out our <a href="https://2014.igem.org/Team:EPF_Lausanne/Envelope_stress_responsive_bacteria">Envelope Stress Responsive Bacteria page !</a></p> | ||
| + | <br /> | ||
| + | |||
| + | --> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | <div class="cntr"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/4/41/Cpx_pathway_description_EPFL.jpg" width="70%" alt="Cpx_pathway_description"> | ||
| + | </div> | ||
| + | --> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <hr /> | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | <h2 class="section-heading" id="howitworks">How the BioPad works - <i>E. coli</i></h2> | ||
| + | |||
| + | <div class="pull-right img-right"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/b/b7/Puce_petit.jpg" data-lightbox="image-0" data-title="A potential BioPad"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/b/b7/Puce_petit.jpg" alt="Potential biopad" height="200" class="img-border" /></a> | ||
| + | <figcaption class="cntr">A potential BioPad</figcaption> | ||
| + | </div> | ||
| + | |||
| + | <p class="lead"> | ||
| + | Our BioPad is a self-designed PDMS microfluidic chip, made of thousands of compartments representing “pixels”. Each 300μm x 300μm x 50μm compartment contains a few layers of <i>E. coli</i>. When the surface of the chip is touched, a deformation on the chip - and thus of the compartments – leads to cellular membrane shear stress and protein aggregation/misfolding in the periplasm.<br/> <br/></p> | ||
| + | |||
| + | |||
| + | <p class="lead"> | ||
| + | |||
| + | The histidine kinase sensor CpxA auto-phosphorylates and transfers its phosphate to its corresponding relay protein, CpxR, resulting in its dimerization. We engineered the pathway, by fusing split reporter protein fragments to the CpxR (IFP1.4). This allows the two fragments to remain inactive until physical interaction of CpxR (stimulated by envelope stress) leads to the proper folding of IFP1.4 and reconstitution of its fluorescent properties. As the reconstitution of the split fragments of IFP1.4 are reversible, the system can be shutdown upon stress removal (CpxA changes conformation to become a phosphatase and induces CpxR’s dissociation).<br/> <br/> | ||
| + | |||
| + | The BioPad also includes a signal detector. This detector is composed of an inexpensive credit card-sized single-board computer called Raspberry Pi, a highly sensitive digital camera with appropriate light filters, and a light-emitting source. It identifies and processes the position of the light/fluorescence emitted by the BioPad. The information about the position of the light relative to chip is then used to control the associated electronic device. <br/> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="cntr"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/1/15/Screen_Shot_2014-10-12_at_3.29.30_PM.png" data-lightbox="image-0" data-title="Fluorescence"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/1/15/Screen_Shot_2014-10-12_at_3.29.30_PM.png" alt="touch bacteria" height="200" class="img-border" /></a> | ||
| + | </div> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <!--<hr /> | ||
| + | <br /> | ||
| + | |||
| + | <h2 id="thehogpathway">The HOG Pathway</h2> | ||
| + | |||
| + | |||
| + | |||
| + | <p class="lead">The HOG (High Osmolarity Glycerol) pathway is a MAPK (Mitogen activated protein kinase) pathway which yeast cells use to coordinate intracellular activities to optimise survival and proliferation in not only hyper-osmotic stress but also heat shock, nitrogen stress and oxidative stress. It is represented below.</p> | ||
| + | |||
| + | <br /> | ||
| + | <br /> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/6/6d/Hog_pathway_copy.jpg" width="750" alt="HOG_pathway_description"> | ||
| + | <br /> | ||
| + | <p> The pathway includes five main proteins:</p> | ||
| + | <ul style="padding-left:80px"> | ||
| + | <li><p class="lead">Sho1/Sln1 – Membrane proteins which are classed as STREs (STress Response Elements) which sense the stress and initiate the pathway</p></li> | ||
| + | <li><p class="lead">Ste11 – The MAPKKK which phosphorylates PBS2</p></li> | ||
| + | <li><p class="lead">PBS2 – The MAPKK which phosphorylates HOG1</p></li> | ||
| + | <li><p class="lead">HOG1 – The MAPK which localizes to the nucleus upon phosphorylation and induces target gene transcription</p></li> | ||
| + | </ul> | ||
| + | <br>--> | ||
| + | <br> | ||
| + | |||
| + | <h2 id="howweengineered">Engineering the HOG pathway in <i>S. cerevisiae</i></h2> | ||
| + | |||
| + | |||
| + | |||
| + | <p class="lead"> | ||
| + | <br/> | ||
| + | The <a target="_blank" href="https://2014.igem.org/Team:EPF_Lausanne/Yeast">HOG pathway</a> controls the response to osmotic stress in yeast cells. | ||
| + | Like bacteria, our modified <i>S. cerevisiae</i> cells can be loaded into the PDMS microfluidic chips we have designed. Thus cells are confined in small compartments made of a soft material. When the surface of the chip is touched, it leads to a deformation of the chip and its chambers. Since the HOG pathway is reactive to turgor pressure, the mechanical pressure applied activates it. Upon induction of the pathway, which is a classical MAP kinase pathway, PBS2 – a MAPKK – is phosphorylated and binds HOG1 – a MAPK – and in turn phosphorylates it. | ||
| + | </p> | ||
| + | |||
| + | <div class="pull-left img-left" style="width:55%"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/7/7b/Schema_split.png" data-lightbox="img1" data-title="Figure of Hog1/Pbs2 split sfGFP and split rLuc"> | ||
| + | <img src="https://static.igem.org/mediawiki/2014/7/7b/Schema_split.png" alt="" class="img-border" style="width: 100%"> | ||
| + | </a> | ||
| + | <figcaption class="cntr">Figure of Hog1/Pbs2 split sfGFP and split rLuc</figcaption> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <p class="lead"> | ||
| + | These two kinases were fused to split fluorescent and luminescent proteins, via a 13-amino acid flexible linker, by homologous recombination. This allowed us to detect the phosphorylation of Hog1 by Pbs2 in response to osmotic pressure or touch. We used split sfGFP and split Renilla luciferase tags on the C-terminals of both proteins. | ||
| + | </p> | ||
| + | |||
| + | <p class="lead"> | ||
| + | The split sfGFP is irreversible and aimed to show the interaction between our two Pbs2 and Hog1 while we used the reversible split luciferase tags to assess the activation and inactivation of the pathway. In fact, when stress is removed, the signal should decline. The BioPad Detector is programmed to identify and process the light position and can transmit the information to a computer. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <hr> | ||
| + | |||
| + | |||
| + | <h2 class="section-heading" id="thebiopad">The BioPad Detector</h2> | ||
| + | <p class="lead"> | ||
| + | |||
| + | |||
| + | <!-- ENGINEERING DETECTOR --> | ||
| + | |||
| + | The signals induced by the BioPad are then processed by our self designed detection system: the BioPad Detector. The BioPad Detector is mainly made of a cheap computer (Raspberry Pi), a highly sensitive digital camera with appropriate light filters, and a light emitting source. The BioPad Detector locates signals from various sources (infrared fluorescence, green fluorescence and luminescence), processes them and sends back the relative positions of the signals with respect to the BioPad. Thanks to this position, we are able to extract information such as giving a computer operating system that the position represents the position of the mouse on a screen, that the well at the given position is a suitable antibiotic candidate, or that a gene of interest has been activated. We therefore effectively control a computer or any other electronic device through a living interface: the BioPad. | ||
| + | |||
| + | </p> | ||
| + | |||
| + | <div class="cntr"> | ||
| + | <a href="https://static.igem.org/mediawiki/2014/b/bd/IMG_4154.JPG" data-lightbox="raspberry" data-title="the Raspberry Pi with a microfluidic chip on the lens"><img src="https://static.igem.org/mediawiki/2014/8/88/DeviceRasp.png" alt="Device" class="img-border img-responsive" /></a> | ||
| + | <figcaption class="cntr">the Raspberry Pi with a microfluidic chip on the lens</figcaption> | ||
| + | </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <!-- END PROJECT --> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | |||
| + | <div class="col col-md-3"> | ||
| + | <nav id="affix-nav" class="sidebar hidden-sm hidden-xs"> | ||
| + | <ul class="nav sidenav box" data-spy="affix" data-offset-top="200" data-offset-bottom="600"> | ||
| + | <li class="active"><a href="#title_intro">Introduction</a></li> | ||
| + | <li><a href="#howitworks">How the BioPad works - <i>E. coli</i></a></li> | ||
| + | <li><a href="#howweengineered">Engineering the HOG pathway in <i>S. cerevisiae</i></a> </li> | ||
| + | <li><a href="#thebiopad">The BioPad Detector</a></li> | ||
| + | |||
| + | </ul> | ||
| + | </nav> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | |||
<!-- END ABSTRACT --> | <!-- END ABSTRACT --> | ||
| + | |||
</html> | </html> | ||
{{CSS/EPFL_bottom}} | {{CSS/EPFL_bottom}} | ||
Latest revision as of 03:22, 18 October 2014
Overview
Introduction
Biological responses can be quick and precise! This is the message that our team wants to convey. With this in mind, our team brought forward a novel idea:
Combining Protein Complementation techniques with biosensors and microfluidics allows fast spatiotemporal analysis of bacterial/yeast responses to stimuli.
Our team explored this hypothesis by engineering two stress related pathways in E. coli and S. cerevisiae with in mind the development of a BioPad: a biological touchscreen consisting of a microfluidic chip, touch-responsive bacteria/yeast, and a signal detector. Learn more about how the BioPad works !
The pathway engineered in E. coli, the Cpx Pathway, is a two-component regulatory system responsive to envelope stress. A full description of the pathway is available here.
In S. cerevisiae we modified the HOG Pathway - a MAPKK pathway responsive to osmotic stress. For more information concerning the HOG Pathway click here.
Our project also includes an extensive microfluidics section. Our self designed chips helped us improve precision, safety and quantification methods used throughout the project. To learn more about the microfluidic components of our project check out this link.
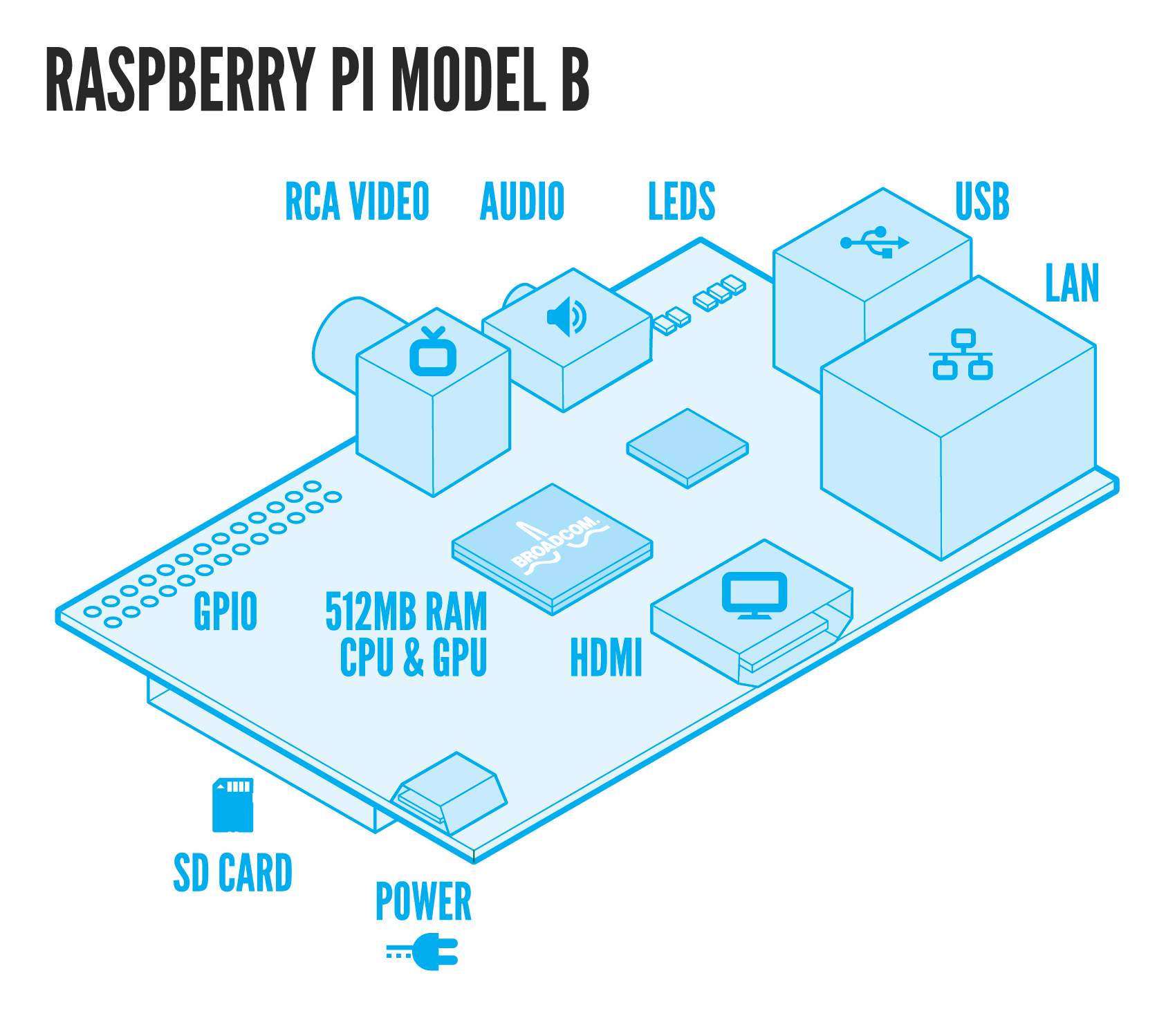
Last but not least, we designed a novel signal detector! To make signal detection more practical we developed an automatised cheap tracking system made of a mini-computer (Raspberry Pi) and a mini-HD camera. More details concerning the BioPad detector can be found here.
How the BioPad works - E. coli
Our BioPad is a self-designed PDMS microfluidic chip, made of thousands of compartments representing “pixels”. Each 300μm x 300μm x 50μm compartment contains a few layers of E. coli. When the surface of the chip is touched, a deformation on the chip - and thus of the compartments – leads to cellular membrane shear stress and protein aggregation/misfolding in the periplasm.
The histidine kinase sensor CpxA auto-phosphorylates and transfers its phosphate to its corresponding relay protein, CpxR, resulting in its dimerization. We engineered the pathway, by fusing split reporter protein fragments to the CpxR (IFP1.4). This allows the two fragments to remain inactive until physical interaction of CpxR (stimulated by envelope stress) leads to the proper folding of IFP1.4 and reconstitution of its fluorescent properties. As the reconstitution of the split fragments of IFP1.4 are reversible, the system can be shutdown upon stress removal (CpxA changes conformation to become a phosphatase and induces CpxR’s dissociation).
The BioPad also includes a signal detector. This detector is composed of an inexpensive credit card-sized single-board computer called Raspberry Pi, a highly sensitive digital camera with appropriate light filters, and a light-emitting source. It identifies and processes the position of the light/fluorescence emitted by the BioPad. The information about the position of the light relative to chip is then used to control the associated electronic device.
Engineering the HOG pathway in S. cerevisiae
The HOG pathway controls the response to osmotic stress in yeast cells.
Like bacteria, our modified S. cerevisiae cells can be loaded into the PDMS microfluidic chips we have designed. Thus cells are confined in small compartments made of a soft material. When the surface of the chip is touched, it leads to a deformation of the chip and its chambers. Since the HOG pathway is reactive to turgor pressure, the mechanical pressure applied activates it. Upon induction of the pathway, which is a classical MAP kinase pathway, PBS2 – a MAPKK – is phosphorylated and binds HOG1 – a MAPK – and in turn phosphorylates it.
These two kinases were fused to split fluorescent and luminescent proteins, via a 13-amino acid flexible linker, by homologous recombination. This allowed us to detect the phosphorylation of Hog1 by Pbs2 in response to osmotic pressure or touch. We used split sfGFP and split Renilla luciferase tags on the C-terminals of both proteins.
The split sfGFP is irreversible and aimed to show the interaction between our two Pbs2 and Hog1 while we used the reversible split luciferase tags to assess the activation and inactivation of the pathway. In fact, when stress is removed, the signal should decline. The BioPad Detector is programmed to identify and process the light position and can transmit the information to a computer.
The BioPad Detector
The signals induced by the BioPad are then processed by our self designed detection system: the BioPad Detector. The BioPad Detector is mainly made of a cheap computer (Raspberry Pi), a highly sensitive digital camera with appropriate light filters, and a light emitting source. The BioPad Detector locates signals from various sources (infrared fluorescence, green fluorescence and luminescence), processes them and sends back the relative positions of the signals with respect to the BioPad. Thanks to this position, we are able to extract information such as giving a computer operating system that the position represents the position of the mouse on a screen, that the well at the given position is a suitable antibiotic candidate, or that a gene of interest has been activated. We therefore effectively control a computer or any other electronic device through a living interface: the BioPad.
 "
"












