Team:CU-Boulder/Results
From 2014.igem.org
Contents |
Our Final System: CRISPR-Cas9 phage
The final System is comprised of two parts: the delivery of the CRISPR-Cas9 machinery and the targeting of the endonuclease.
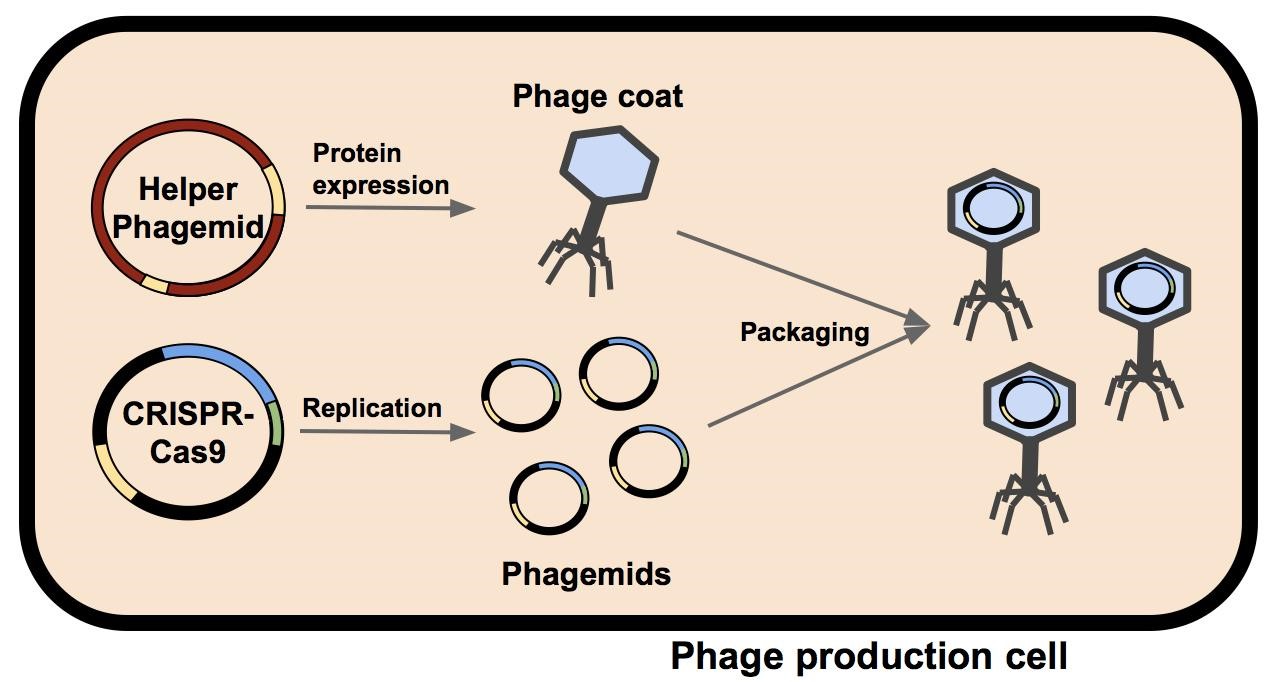
M13 phage were used as the delivery mode in this system. Depicted above is a single bacterium containing a helper phagemid and a phagemid. The phage coat and assembly proteins are expressed from the helper phagemid but this packaging signal was disrupted by the insertion of a selectable marker (either kanamycin in M13K07 or ampicillin in M13g6A1, see below) and a plasmid origin of replication so its ability to be uptaken into a phage capsid is hindered. The phagemid contains the construct to be delivered (in this case, the CRISPR-Cas9 construct) and a fully functional packaging signal so it will be packaged into the assembling phage. The produced phage containing the phagemid are isolated and can be used to infect a target population of bactium.
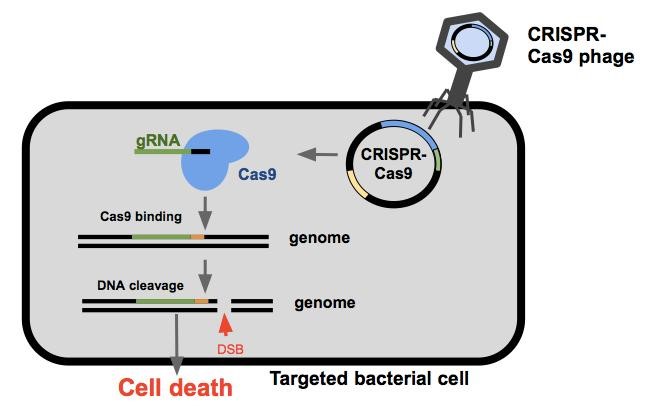
Once the CRISPR-Cas9 phagemid enters the cell, the bacterium will express the Cas9 endonuclease and a guide RNA (gRNA, target sequence is determined by spacer). These two parts come together and search the bacterial genome for PAM sites (Protospacer Adjacent Motifs, shown here is orange). The Cas9 will bind to the PAM site allowing the gRNA to anneal to the target sequence (shown here in green). If binding is successful, the Cas9 endonuclease cleaves the DNA resulting in a double stranded break. If the cell is unable to repair itself or does so incorrectly, the cell will die.
New BioBricked Parts
- BBa_K1445000: The M13 origin of replication (M13ori) is a noncoding sequence that when cloned onto a plasmid, facilitates the uptake of that plasmid into an M13 phage capsid.
- BBa_K1445001: M13ori (BBa_K1445000) cloned onto a CRISPR-Cas9 construct (BBa_K1218011). The CRISPR-Cas9 part codes for a Cas9 endonuclease that is guided to a DNA sequence specified by a spacer located within the minimal CRISPR array. Inclusion of the M13ori allows for its uptake into M13 phage.
- BBa_K1445002: BioBricked phagemid vector allows for the easy cloning of BioBrick parts into a backbone that can be packaged into a phage without the addition of the M13ori to the insert region.
- Additionally, the genes for M13 phage coat proteins were BioBricked and cloned onto a pSB6A1 vector. This allows the phage producing genes to be cloned onto vectors with a different resistance marker if the resistance interferes with the marker on the phagemid. This part was not submitted to the registry.
Phage delivery system (M13ori)
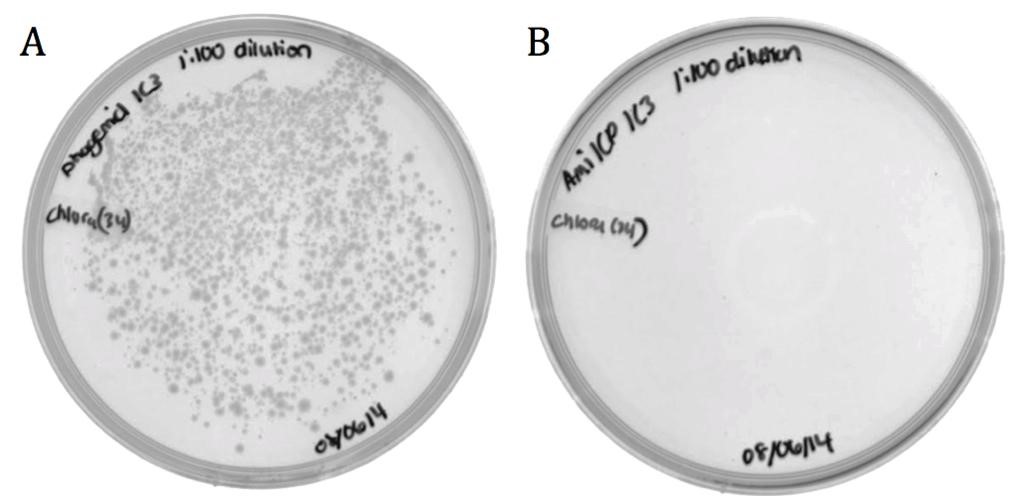
The M13 origin of replication (M13ori) has been documented as the packaging signal for the M13 phage. When single stranded, the M13ori forms unique hairpins that signal packaging into a phage capsid. To verify its functionality, its packaging efficiency was compared to that of amilCP. The amilCP sequence is roughly equal in length to the M13ori and both parts were cloned onto the pSB1C3 backbone; therefore, the nucleotide sequence is the only distinguishing factor. The helper phagemid M13K07 (a plasmid containing M13 phage structural proteins and a weakened packaging signal) made phage in host cells containing pSB1C3-M13ori or pSB1C3-amilCP phagemids. ER2738 E. coli were infected by phage from the M13ori or amilCP sample to assess the respective packaging efficiencies.
Figure 1: Conjugated BW23115 E. coli infected with recombinant phage A) pSB1C3-M13ori or B) pSB1C3-amilCP phagemids on LB agar with 170 ug/mL chloramphenicol.
The growth in the M13ori sample demonstrates that this phagemid was packaged and delivered by the M13 phage. The absence of growth in the amilCP sample proves that without the presence of a packaging signal, a plasmid cannot be taken up by a phage capsid. These results show that the M13ori is necessary and sufficient for phagemid packaging.
Making phage with M13g6A1
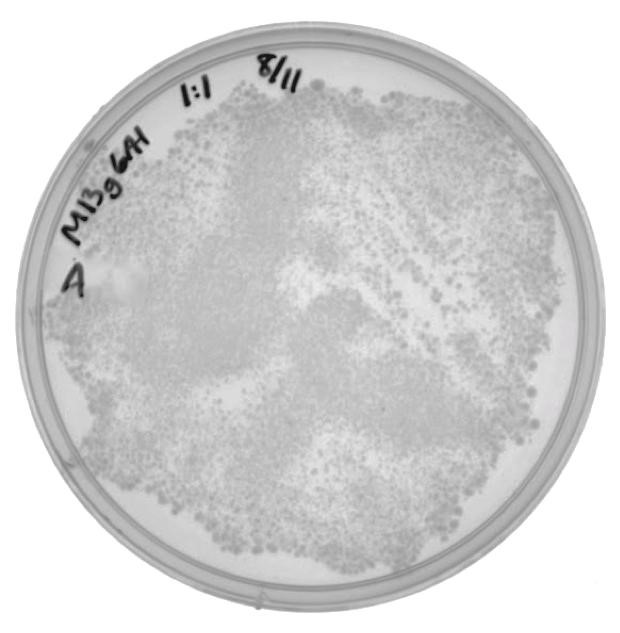
For the previously mentioned experiment, M13K07 was used as the helper phagemid. However, this part contains a kanamycin resistance gene, which may interfere with the analysis of Cas9 targeting of the neomycin phosphotransferase gene, which also confers kanamycin resistance. The genes that code for the phage coat proteins were amplified from M13K07 and cloned onto the pSB6A1 vector. Phage amplification using M13g6A1 as the helper phagemid (and no additional phagemid) produced M13 phage packaging M13g6A1. ER2738 cells were infected with the isolated phage and were plated onto ampicillin to verify the viability of phage produced by this helper phagemid.
Figure 2. The presence of colonies in Figure 2 confirm that M13g6A1 is capable of producing viable phage.
Program to design a Spacer Sequence
For the following Cas9 experiments, a spacer sequence was designed to target the neomycin phosphotransferase gene present in the targeting strain. This program inputs FASTA files in two sets. In one are the sequences that should be targeted for Cas9 binding while the other set consists of sequences that should not be targeted. It then searches for possible targeting sites that are present in the targeting sequences and absent from the sequences that should not be targeted. The best spacer sequences are then presented to the user.
For this project, the sequence for the neomycin phosphotransferase gene was given to the program as the targeting sequence. The E. coli K-12 and E. coli MG1655 genomes were given to the program as sequences to no target. One of the output spacers was chosen for use in the following CRISPR-Cas9 experiments.
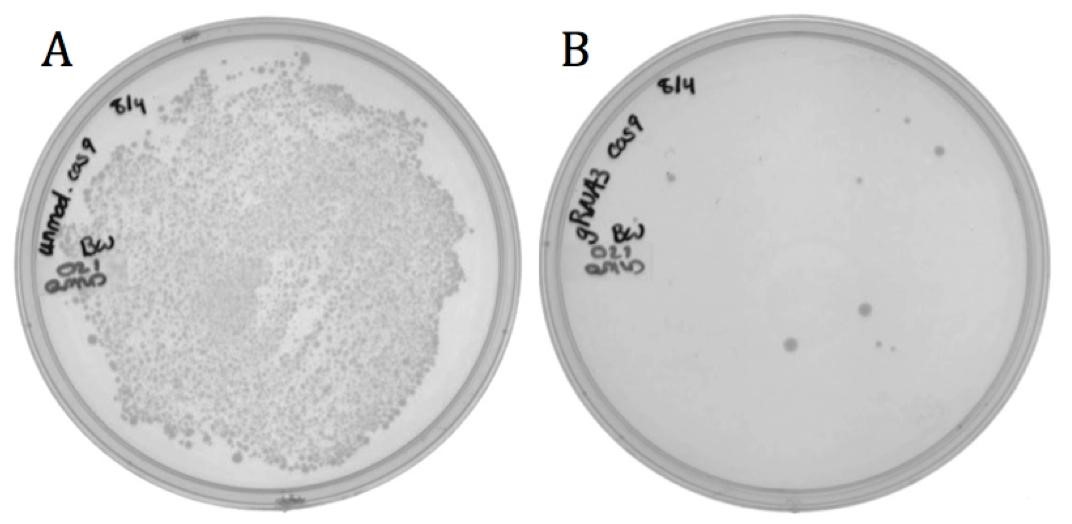
Targeted killing of BW through Transformation (characterization of BBa_K1218011) The spacer designed to target the neomycin phosphotransferase gene was cloned into the CRISPR-Cas9 construct (BBa_K1218011). The unmodified spacer does not target the production or target strain so was used as a non-targeting control. The targeting and non-targeting constructs were transformed into the BW23115 E. coli containing the neomycin phosphotransferase through the heat shock method and selected for on chloramphenicol.
Figure 3: Transformation results of neomycin resistant E. coli with Cas9 part having either A) non-targeting or B) targeting spacer sequence on LB agar with 170 ug/mL chloramphenicol.
The decrease in growth between the non-targeting (1920 colonies) and the targeting (8 colonies) samples is accredited to the difference in spacer sequence. When the spacer sequence exists in the genome of cell, the Cas9 endonuclease is able to bind and cleave the DNA, resulting in cell death, making it possible to intentionally kill bacteria by sequence specific Cas9 targeting through transformation.
Interestingly, some targeted colonies survived as can be seen in Figure 1B. Sequencing showed that all eight colonies had deleted the spacer region and one or both of the adjacent repeats.
Cas9 delivery through phage
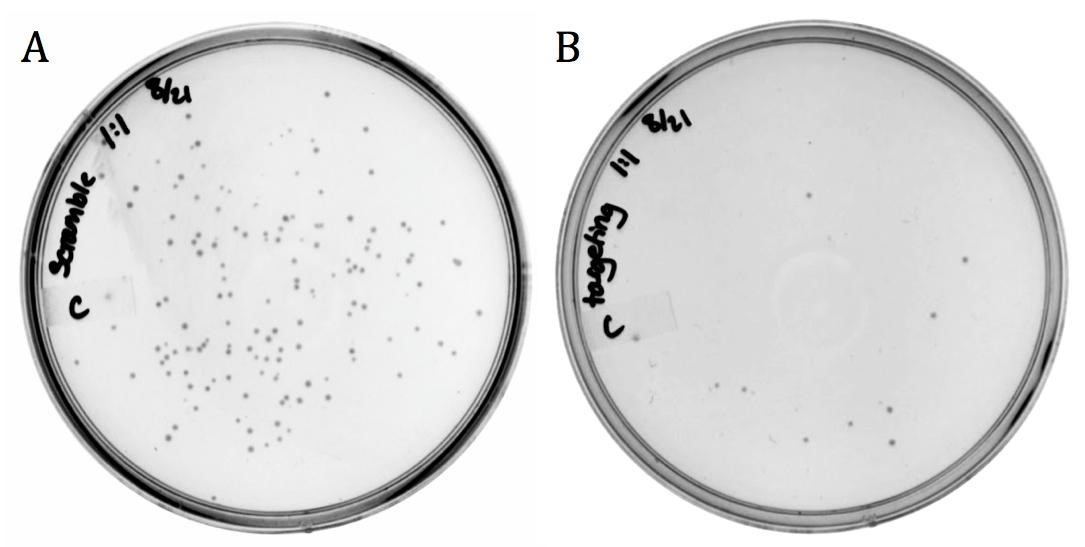
The M13 origin of replication, or M13ori (BBa_K1445000) was cloned upstream of the CRISPR-Cas9 construct (BBa_ K1218011) in the targeting and the non-targeting varieties to allow for their uptake into M13 phage. This composite part was packaged into phage using the M13g6A1 helper phagemid mentioned above. Progeny phage infected conjugated versions of the BW23115 E. coli containing the neomycin phosphotransferase gene and cells were plated onto chloramphenicol.
Figure 4: Infection results of M13 phage delivering A) non-targeting and B) targeting varieties of part BBa_K1445001 on LB agar with 170 ug/mL chloramphenicol.
The presence of growth in both samples demonstrates that the addition of the M13ori does allow for packaging and delivery by the M13 phage. The decrease in growth from the non-targeting (143 colonies) to the targeting (11 colonies) is accredited to the differences in spacer sequence. With a spacer sequence that targets within the genome of the cell, the Cas9 endonuclease is able to bind and cleave the DNA, successfully killing the bacterium.
Unmodified spacer may have off-target binding to FҠepisome, causing cell death in the production strain or targeting strain. (I tried but could NOT find the sequence. Any idea how we can say we donӴ know without sounding lazy?)
Additional Work and Characterization (BBa_K1445002)
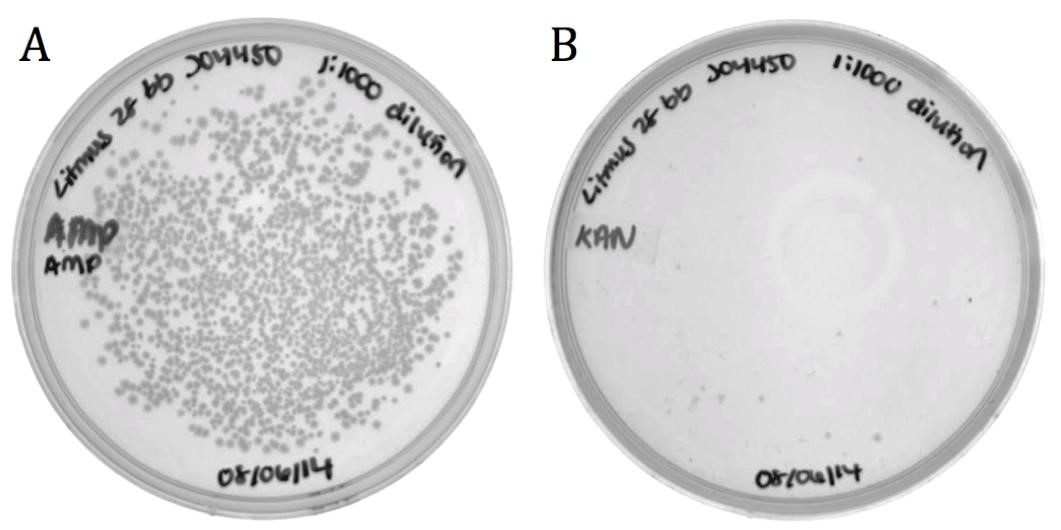
Part BBa_J04450 and flanking regions to the sequencing primer sites was cloned into the Litmus28i vector to make a biobricked phagemid backbone that can be packaged into M13 phage. To verify functionality, the vector was packaged into phage using the M13K07 helper phagemid. Progeny phage were infected ER2738 e. coli and cells were selected for on ampicillin.
Figure 5: Packaging efficiency of recombinant phagemid to helper phagemid packaging. A) Sample diluted 1:1000 and grown on 100 ug/mL Ampicillin to select for BBa_K1445002. B) Sample diluted 1:1000 and grown on 50 ug/mL Kanamycin to select for M13K07 helper phagemid. BBa_K1445002 was packaged over M13K07 at a ratio of 136:1.
The growth on ampicillin (1496 colonies) shows that the biobricked version of Litmus28i retains its ability to be packaged and delivered by M13 phage. Additionally, the same sample was plated on kanamycin to determine the packaging ratio of phagemid to helper phagemid. As shown in Figure 1B), very little growth was present on kanamycin (11 colonies) confirming that BBa_K1445002 was preferentially packaged over the M13K07 helper phagemid at a ratio of 136:1.
 "
"