Team:CU-Boulder/Notebook/Protocols
From 2014.igem.org
Jimmynovik (Talk | contribs) |
|||
| (58 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
__FORCETOC__ | __FORCETOC__ | ||
| - | ==Amplification | + | ==Phage Amplification== |
'''Need''' | '''Need''' | ||
| - | *Plate of infectable cells that contain F’ episome | + | :*Plate of infectable cells that contain F’ episome |
| - | *2.5M NaCl/20% PEG-8000 | + | :*2.5M NaCl/20% PEG-8000 |
| - | *1x TBS | + | :*1x TBS |
'''Day 1''' | '''Day 1''' | ||
| - | + | :1.Add a fresh colony of infectable cells to 50mL LB in 125mL flask. | |
| - | + | ::a. Include phagemid antibiotic only. | |
| - | + | ::b. Grow at 37°C, 250rpm until OD is between 0.03 and 0.05 | |
| - | + | :2.Add the helper phage to a final concentration of 1 x10^8 phage/mL | |
| - | + | :3.Incubate for 60-90 minutes, shaking | |
| - | + | :4.Add Helper Phagemid antibiotic to a high concentration | |
| - | + | :5.Grow for 14-18 hours at 37°C, shaking | |
'''Day 2''' | '''Day 2''' | ||
| Line 36: | Line 36: | ||
#Spin briefly. Remove supernatant with pipet | #Spin briefly. Remove supernatant with pipet | ||
#Resuspend pellet in 200ul 1x TBS. | #Resuspend pellet in 200ul 1x TBS. | ||
| - | + | ::a. If desired, combine contents of both tubes into one | |
| + | <br> | ||
| + | ''Protocol from “Use of M13KO7 Helper Phage for isolation of single-stranded phagemid DNA” by NEB'' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| - | + | ==Heat Shock Transformation== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
'''Before you start''' | '''Before you start''' | ||
| - | *Heat hot plate or water bath to 42°C | + | :*Heat hot plate or water bath to 42°C |
| - | *Warm selection plates to 37°C | + | :*Warm selection plates to 37°C |
'''Transformation''' | '''Transformation''' | ||
| - | + | :1.Thaw chemically competent cells on ice for 10-15 minutes | |
| - | + | :2.Add 40ul cells to fresh 1.7mL tube | |
| - | + | :3.Add DNA | |
| - | + | ::a. If using a ligation product add up to 10ul of sample | |
| - | + | ::b. If using supercoiled plasmid add 100ng | |
| - | + | :4.Incubate on ice for 30 minutes | |
| - | + | :5.Heat shock cells on hot plate (or water bath) for 30-45s* @ 42°C | |
| - | + | :6.Incubate on ice for 2-5 minutes | |
| - | + | :7.Add 200 μL SOC and shake gently for 1-2 hours @ 37° C | |
| - | + | ::a.('''Note''': Can also recover in 960ul. After recovery, gently spin cells and remove supernatant. Resuspend in 200ul LB) | |
| - | + | :8.Plate 100-200ul cells onto selection plates | |
| - | + | ::a. If high efficiency is expected, we suggest also plating a 1:10 dilution | |
| - | + | :9.Once dry, turn upside down (agar on top) and incubate overnight @ 37° C | |
| - | Optimal timing depends on cells | + | ::*Optimal timing depends on cells |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Bacterial Conjugation== | ==Bacterial Conjugation== | ||
'''Need''' | '''Need''' | ||
| - | *Donor cells: Cells already containing F’ episome | + | :*Donor cells: Cells already containing F’ episome |
| - | *Recipient cells: Cells with a resistance marker that is absent from donor cells | + | :*Recipient cells: Cells with a resistance marker that is absent from donor cells |
| - | *Double selection plate containing antibiotic to select for F’ episome and a second antibiotic to select for the recipient cells | + | :*Double selection plate containing antibiotic to select for F’ episome and a second antibiotic to select for the recipient cells |
'''Day 1''' | '''Day 1''' | ||
| - | #Set up liquid overnight of donor cells. Include antibiotic | + | #Set up liquid overnight of '''donor''' cells. Include antibiotic |
| - | #Set up liquid overnight of recipient cells. Include antibiotic | + | #Set up liquid overnight of '''recipient''' cells. Include antibiotic |
'''Day 2''' | '''Day 2''' | ||
| - | + | :1.Mix 500ul of each overnight sample in a new tube. Mix by pipetting or flicking | |
| - | + | :2.Incubate for 30 minutes at 37°C, shaking | |
| - | + | :3.Plate 100ul onto double selection plate | |
| - | + | ::a.We advise also plating a 1:10 dilution | |
| - | #Incubate at 37°C | + | :4.Incubate at 37°C |
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==CaCl2 Competent Cells== | ||
| + | |||
| + | '''To prepare for Day 2''' | ||
| + | :* Set centrifuge to 4°C | ||
| + | :* MgCl2 and CaCl2 solutions | ||
| + | :* Thaw DMSO | ||
| + | :* Chill tubes | ||
| + | :* Acquire liquid nitrogen or dry ice | ||
| + | |||
| + | '''Day 1''' | ||
| + | #Grow cells O/N | ||
| + | |||
| + | '''Day 2''' | ||
| + | :1.Add 0.5mL of the overnight culture to 50mL LB | ||
| + | :2.Grow until OD is between 0.2 and 0.4 | ||
| + | :3.Incubate on ice for 30 minutes | ||
| + | :4.Centrifuge for 10 minutes at 2700 x g and 4°C | ||
| + | :5.Decant. Dry upside down on a paper towel for 1 minute | ||
| + | :6.Completely resuspend in 30mL 0.8M MgCl2, 0.2M CaCl2 | ||
| + | ::a. Gently vortex | ||
| + | :7.Centrifuge for 10 minutes at 2700 x g and 4°C | ||
| + | :8.Decant. Dry upside down on a paper towel for 1 minute | ||
| + | :9.Fully resuspend in 2mL of 0.1M CaCl2 | ||
| + | :10.Chill sample on ice. Add 70ul DMSO, keeping the sample tube on ice | ||
| + | :11.Swirl to mix | ||
| + | :12.Incubate on ice for 15 minutes | ||
| + | :13.Add 70ul DMSO, swirl to mix, keeping the sample tube on ice | ||
| + | :14.Dispense 200ul into pre-chilled 1.7mL tubes | ||
| + | :15.Snap freeze with liquid nitrogen or dry ice | ||
| + | :16.Store at -80°C until ready to use | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==Phagemid DNA Isolation== | ||
| + | |||
| + | |||
| + | '''Need''' | ||
| + | |||
| + | :*Fresh plate of infectable cells (contain F’ episome) | ||
| + | :*2.5M NaCl/20% PEG-8000 | ||
| + | :*TBS, TE, phenol, phenol/chloroform, chloroform | ||
| + | |||
| + | '''Day 1''' | ||
| + | |||
| + | :1.Add a fresh colony to 50mL LB in 125mL flask. Grow at 37°C, 250rpm until OD is between 0.03 and 0.05 | ||
| + | :2.Add M13KO7 helper phage to a final concentration of 1 x10^8 phage/mL* | ||
| + | :3.Continue shaking for 60-90 minutes | ||
| + | :4.Add Kanamycin to final concentration of 70ug/mL | ||
| + | :5.Grow for 14-18 hours at 37°C, 250rpm | ||
| + | |||
| + | :*Can use a different helper phage if needed. In step 4, add the antibiotic specific to the Helper Phagemid | ||
| + | |||
| + | '''Day 2''' | ||
| + | |||
| + | :1.Spin culture at 4,000 x g for 10 minutes | ||
| + | :2.Transfer supernatant to a fresh conical. Repeat spin on supernatant | ||
| + | :3.Transfer the upper 90% of supernatant to a new conical | ||
| + | :4.Add 0.2 volume of 2.5M NaCl/20% PEG-8000 to the new conical. Gently mix | ||
| + | :5.Incubate at 4°C for at least 60 minutes | ||
| + | :6.Centrifuge at 12,000 x g for 10 minutes. Carefully decant | ||
| + | :7.Spin again briefly | ||
| + | :8.Gently resuspend pellet in 1.6mL 1x TBS | ||
| + | :9.Aliquot 800ul into 2 microfuge tubes. Proceed with both tubes | ||
| + | :10.Spin sample for 1 minute to pellet remaining cells. Transfer supernatant to fresh tubes | ||
| + | :11.Add 160ul of 2.5M NaCl/20% PEG-8000 solution to each | ||
| + | :12.Let sit at room temperature for 5 minutes | ||
| + | :13.Spin at 1300 x g for 10 minutes | ||
| + | :14.Decant the supernatant | ||
| + | :15.Spin briefly. Remove supernatant with pipet | ||
| + | :16.Resuspend pellets in 300ul TE | ||
| + | :17.Phenol extraction: add 300ul phenol. Vortex for 15 seconds | ||
| + | ::a.Let sit for 15 minutes. Spin for 10 minutes | ||
| + | :18.Add H2O so volume samples is about 300ul | ||
| + | :19.Phenol/chloroform extraction*: add 300ul PCIA | ||
| + | ::a.Vortex for 15 seconds. Spin for 10 minutes | ||
| + | :20.Repeat Phenol/Chloroform extraction | ||
| + | :21.Chloroform extraction*: add 300ul chloroform | ||
| + | ::a.Vortex 15 seconds. Spin for 10 minutes | ||
| + | :22.Add 30ul 2.5M NaAc (pH 4.8) | ||
| + | :23.Add 2-2.5 volumes ethanol | ||
| + | :24.Let precipitate for ~2 hours at -20°C | ||
| + | :25.Spin for 1 minute | ||
| + | :26.Decant supernatant | ||
| + | :27.Resuspend in 25-50ul TE | ||
| + | |||
| + | :*Performing steps at 4°C helps with separation | ||
| + | <br> | ||
| + | ''Protocol from: “Use of M13K07 Helper Phage for isolation of single-stranded phagemid DNA” by NEB'' | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==M13 Amplification== | ||
| + | |||
| + | This protocol is to make more M13 phages. | ||
| + | |||
| + | '''Need''' | ||
| + | |||
| + | :*Fresh plate of infectable cells (contain F’ episome) | ||
| + | |||
| + | '''Day 1''' | ||
| + | :1.Grow liquid overnight culture of infectable cells | ||
| + | |||
| + | '''Day 2''' | ||
| + | |||
| + | :1.Add 200ul overnight culture to 20mL LB in a 250mL flask | ||
| + | :2.Add 1ul phage suspension | ||
| + | :3.Incubate for 4-5 hours at 37°C, 250rpm | ||
| + | :4.Centrifuge for 10 minutes at 4500 x g | ||
| + | :5.Transfer supernatant to a new tube. | ||
| + | :6.Repeat centrifugation on supernatant | ||
| + | :7.Transfer top 16mL of supernatant to a new tube | ||
| + | :8.Add 4mL of 2.5M NaCl/20% PEG-8000. Briefly mix | ||
| + | :9.Precipitate phage for 1 hour or overnight at 4°C | ||
| + | :10.Centrifuge for 15 minutes at 12000 x g. Decant supernatant | ||
| + | ::a. Spin briefly. Remove residual supernatant with pipet | ||
| + | :11.Resuspend pellet in 1mL 1x TBS. Transfer to 1.7mL tube | ||
| + | :12.Spin briefly to remove cell debris | ||
| + | :13.Transfer supernatant to a new tube | ||
| + | :14.Add 200ul of 2.5M NaCl/20% PEG-8000 | ||
| + | :15.Incubate on ice for 15-60 minutes | ||
| + | :16.Spin for 10 minutes at 12000-14000 rpm. Discard supernatant | ||
| + | :17.Briefly spin. Remove supernatant with pipette | ||
| + | :18.Resuspend pellet in 200uL TBS | ||
| + | :19.For long term storage at -20C, add 200uL glycerol | ||
| + | |||
| + | |||
| + | ''Adapted from M13 Amplification protocol from NEB'' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==Mini-prep== | ||
| + | |||
| + | '''Notes before starting''' | ||
| + | |||
| + | : '''Optional:''' Add LyseBlue reagent to Buffer P1 at a ratio of 1 to 1000 | ||
| + | |||
| + | :*Add the provided RNase A solution to Buffer P1, mix, '''store bottle at 2-8°C''' | ||
| + | |||
| + | :*Add ethanol (96-100%) to Buffer PE before use | ||
| + | |||
| + | :*All centrifugation steps are carried out at 13,000 rpm (~17,900xg) in a conventional table-top microcentrifuge | ||
| + | |||
| + | |||
| + | '''Mini-prep''' | ||
| + | :1.Centrifuge 1-6mL bacterial overnight culture at >8000 rpm (6800xg) for 3 minutes at room temperature (15-25C) | ||
| + | :2.Resuspend pellet in 250ul Buffer P1 and transfer to microcentrifuge tube | ||
| + | :3.Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear. | ||
| + | ::a.DO NOT allow lysis reaction to proceed for more than 5 minutes | ||
| + | ::b.If using LyseBlue reagent, the solution will turn blue | ||
| + | :4.Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times. | ||
| + | ::a.If using LyseBlue reagent, the solution will turn colorless | ||
| + | :5.Centrifuge for 10 minutes | ||
| + | :6.Apply supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60 s and discard the flow-through | ||
| + | :7.Recommended: Wash the QIAprep spin column by adding 500 ul Buffer PB. Centrifuge for 30-60 s and discard the flow-through | ||
| + | ::a.Only required when using endA+ strains or other bacterial strains with high nuclease activity or carbohydrate content | ||
| + | :8.Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 30-60 s and discard the flow-through | ||
| + | :9.Centrifuge for 1 minutes to remove residual wash buffer | ||
| + | :10.Place the QIAprep column in a clean 1.5mL microcentrifuge tube. To elute DNA, add 30ul Buffer EB. Let stand for 1 min, and centrifuge for 1 minute | ||
| + | ::a.Can elute DNA in 50ul but this will decrease DNA concentration | ||
| + | ::b.To increase yield, let sit for up to 4 minutes | ||
| + | <br> | ||
| + | ''Protocol and Reagent recipes from QIAGEN unless stated otherwise'' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==Bacterial Infection== | ||
| + | |||
| + | '''Need''' | ||
| + | :* Fresh plate of cells that contain the F’ episome | ||
| + | |||
| + | '''Day 1''' | ||
| + | :1.Pick colony from plate and grow overnight | ||
| + | |||
| + | '''Day 2''' | ||
| + | :1.Dilute overnight to OD600 0.1 | ||
| + | :2.Incubate until OD600 ~ 0.5* | ||
| + | :3.Add phage to a final concentration of 1 x108 phage/mL | ||
| + | :4.Incubate for 30 minutes to infect | ||
| + | :5.Plate samples under selection | ||
| + | ::a.We recommend trying a range of dilutions | ||
| + | |||
| + | |||
| + | :'''Note:''' All incubations are done at 37°C and shaking at 225rpm | ||
| + | :'''Note:''' If multiple samples are to be done in parallel | ||
| + | :*If using the same cells, we suggest growing one large batch of cells. Once the OD has reached ~0.5, divide into 50mL samples then add phage. | ||
| + | :*If using different cells, grow each sample to ~0.5 then store on ice. Once all samples have reached ~0.5, incubate on ice for another 30 minutes. Warm for 20-30 minutes at 37C, 250 rpm. Measure OD again to check that samples are comparable (yes, they will have grown some). Add phage | ||
| + | |||
| + | <br> | ||
| + | ''Protocol from “Eliminating Helper phage from Phage Display'' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | ==Cloning a new spacer into pCas9== | ||
| + | |||
| + | '''[[File:UCB-lab protocol cloning-141010-01.jpg]]''' | ||
| + | |||
| + | '''Design the new spacer''' | ||
| + | |||
| + | '''[[File:UCB-lab protocol cloning-141010-02.jpg]]''' | ||
| + | |||
| + | :1. Find a PAM site (5’- NGG -3’). This will be on the 3’ end of the targeting sequence | ||
| + | :2. The 30bps upstream of NGG motif (on the same strand) will be the spacer | ||
| + | :3. Oligo I (fwd. primer): To the spacer sequence, add AAAC to the 5’ end and G to the 3’ end | ||
| + | ::a. NOTE: These nucleotides are for ligation purposes and are NOT used in targeting. Therefore, they do not have to be present in the target sequence and do not count towards the 30bps | ||
| + | :4. Oligo II (rvs. primer): Find the reverse complement of the spacer sequence. Add CAAAA to the 5’ end. | ||
| + | |||
| + | '''Phosphorylation''' | ||
| + | :1.Mix the following reactants | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !Reactant | ||
| + | !Amount | ||
| + | |- | ||
| + | |Oligo I (100uM) | ||
| + | |1 ul | ||
| + | |- | ||
| + | |Oligo II (100uM) | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10x T4 Ligase Buffer | ||
| + | |5 ul | ||
| + | |- | ||
| + | |T4 PNK | ||
| + | |1 ul | ||
| + | |- | ||
| + | |H2O | ||
| + | |42 ul | ||
| + | |} | ||
| + | |||
| + | :2.Incubate for 60 minutes at 37C then 20 minutes at 65C | ||
| + | |||
| + | '''Anneal Oligos''' | ||
| + | :1. Add 2.5 ul of 1M NaCl to the phosphorylation product | ||
| + | :2. Incubate for 5 minutes at 95C then let slowly cool to room temperature. | ||
| + | ::a. It is recommended to use a thermocycler that either cools at a gradient or steps down in temperature over the course of ~2 hours. | ||
| + | ::b. Alternatively, incubate the sample at 95C for 5 minutes then remove the heat block from the heater and let it cool naturally for 2 hours | ||
| + | :3. Dilute the annealed oligos 1:10 | ||
| + | |||
| + | '''Vector digest with BsaI''' | ||
| + | :1. Mix the following reactants | ||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !Reactant | ||
| + | !Amount | ||
| + | |- | ||
| + | |Cutsmart Buffer | ||
| + | |5 ul | ||
| + | |- | ||
| + | |BsaI enzyme | ||
| + | |1 ul | ||
| + | |- | ||
| + | |DNA | ||
| + | |1-2ug | ||
| + | |- | ||
| + | |H2O | ||
| + | |Bring volume to 50ul | ||
| + | |} | ||
| + | :2. Incubate for 90 minutes at 37C then 20 minutes at 65C | ||
| + | :3. Gel extract digested pCas9 | ||
| + | |||
| + | '''Ligation''' | ||
| + | :1. Mix the following reactants | ||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | !Reactant | ||
| + | !Amount | ||
| + | |- | ||
| + | |Gel extracted vector | ||
| + | |1 ul | ||
| + | |- | ||
| + | |Diluted annealed oligos | ||
| + | |2 ul | ||
| + | |- | ||
| + | |10x T4 Ligase Buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |T4 Ligase | ||
| + | |1 ul | ||
| + | |- | ||
| + | |H2O | ||
| + | |14 ul | ||
| + | |} | ||
| + | :2. Incubate overnight (8-12 hours) at 16C | ||
| + | |||
| + | '''Transform''' | ||
| + | :1. Transform into DH5alpha | ||
| + | |||
| + | |||
| + | ''Protocol from Marraffini Lab'' | ||
| + | |||
| + | |||
| + | |||
| + | |||
| - | |||
{{Template:UCB-Footer}} | {{Template:UCB-Footer}} | ||
Latest revision as of 09:28, 17 October 2014
Contents |
Phage Amplification
Need
- Plate of infectable cells that contain F’ episome
- 2.5M NaCl/20% PEG-8000
- 1x TBS
Day 1
- 1.Add a fresh colony of infectable cells to 50mL LB in 125mL flask.
- a. Include phagemid antibiotic only.
- b. Grow at 37°C, 250rpm until OD is between 0.03 and 0.05
- 2.Add the helper phage to a final concentration of 1 x10^8 phage/mL
- 3.Incubate for 60-90 minutes, shaking
- 4.Add Helper Phagemid antibiotic to a high concentration
- 5.Grow for 14-18 hours at 37°C, shaking
Day 2
- Spin culture at 4,000 x g for 10 minutes
- Transfer supernatant to a fresh conical. Repeat spin on supernatant
- Transfer the upper 90% of supernatant to a new conical
- Add 0.2 volume of 2.5M NaCl/20% PEG-8000 to the new conical. Gently mix
- Incubate at 4°C for at least 60 minutes
- Centrifuge at 12,000 x g for 10 minutes. Carefully decant
- Spin again briefly
- Gently resuspend pellet in 1.6mL 1x TBS
- Aliquot 800ul into 2 microfuge tubes. Proceed with both tubes
- Spin sample for 1 minute to pellet remaining cells. Transfer supernatant to fresh tubes
- Add 160ul of 2.5M NaCl/20% PEG-8000 solution to each
- Let sit at room temperature for 5 minutes
- Spin at 1300 x g for 10 minutes
- Decant the supernatant
- Spin briefly. Remove supernatant with pipet
- Resuspend pellet in 200ul 1x TBS.
- a. If desired, combine contents of both tubes into one
Protocol from “Use of M13KO7 Helper Phage for isolation of single-stranded phagemid DNA” by NEB
Heat Shock Transformation
Before you start
- Heat hot plate or water bath to 42°C
- Warm selection plates to 37°C
Transformation
- 1.Thaw chemically competent cells on ice for 10-15 minutes
- 2.Add 40ul cells to fresh 1.7mL tube
- 3.Add DNA
- a. If using a ligation product add up to 10ul of sample
- b. If using supercoiled plasmid add 100ng
- 4.Incubate on ice for 30 minutes
- 5.Heat shock cells on hot plate (or water bath) for 30-45s* @ 42°C
- 6.Incubate on ice for 2-5 minutes
- 7.Add 200 μL SOC and shake gently for 1-2 hours @ 37° C
- a.(Note: Can also recover in 960ul. After recovery, gently spin cells and remove supernatant. Resuspend in 200ul LB)
- 8.Plate 100-200ul cells onto selection plates
- a. If high efficiency is expected, we suggest also plating a 1:10 dilution
- 9.Once dry, turn upside down (agar on top) and incubate overnight @ 37° C
- Optimal timing depends on cells
Bacterial Conjugation
Need
- Donor cells: Cells already containing F’ episome
- Recipient cells: Cells with a resistance marker that is absent from donor cells
- Double selection plate containing antibiotic to select for F’ episome and a second antibiotic to select for the recipient cells
Day 1
- Set up liquid overnight of donor cells. Include antibiotic
- Set up liquid overnight of recipient cells. Include antibiotic
Day 2
- 1.Mix 500ul of each overnight sample in a new tube. Mix by pipetting or flicking
- 2.Incubate for 30 minutes at 37°C, shaking
- 3.Plate 100ul onto double selection plate
- a.We advise also plating a 1:10 dilution
- 4.Incubate at 37°C
CaCl2 Competent Cells
To prepare for Day 2
- Set centrifuge to 4°C
- MgCl2 and CaCl2 solutions
- Thaw DMSO
- Chill tubes
- Acquire liquid nitrogen or dry ice
Day 1
- Grow cells O/N
Day 2
- 1.Add 0.5mL of the overnight culture to 50mL LB
- 2.Grow until OD is between 0.2 and 0.4
- 3.Incubate on ice for 30 minutes
- 4.Centrifuge for 10 minutes at 2700 x g and 4°C
- 5.Decant. Dry upside down on a paper towel for 1 minute
- 6.Completely resuspend in 30mL 0.8M MgCl2, 0.2M CaCl2
- a. Gently vortex
- 7.Centrifuge for 10 minutes at 2700 x g and 4°C
- 8.Decant. Dry upside down on a paper towel for 1 minute
- 9.Fully resuspend in 2mL of 0.1M CaCl2
- 10.Chill sample on ice. Add 70ul DMSO, keeping the sample tube on ice
- 11.Swirl to mix
- 12.Incubate on ice for 15 minutes
- 13.Add 70ul DMSO, swirl to mix, keeping the sample tube on ice
- 14.Dispense 200ul into pre-chilled 1.7mL tubes
- 15.Snap freeze with liquid nitrogen or dry ice
- 16.Store at -80°C until ready to use
Phagemid DNA Isolation
Need
- Fresh plate of infectable cells (contain F’ episome)
- 2.5M NaCl/20% PEG-8000
- TBS, TE, phenol, phenol/chloroform, chloroform
Day 1
- 1.Add a fresh colony to 50mL LB in 125mL flask. Grow at 37°C, 250rpm until OD is between 0.03 and 0.05
- 2.Add M13KO7 helper phage to a final concentration of 1 x10^8 phage/mL*
- 3.Continue shaking for 60-90 minutes
- 4.Add Kanamycin to final concentration of 70ug/mL
- 5.Grow for 14-18 hours at 37°C, 250rpm
- Can use a different helper phage if needed. In step 4, add the antibiotic specific to the Helper Phagemid
Day 2
- 1.Spin culture at 4,000 x g for 10 minutes
- 2.Transfer supernatant to a fresh conical. Repeat spin on supernatant
- 3.Transfer the upper 90% of supernatant to a new conical
- 4.Add 0.2 volume of 2.5M NaCl/20% PEG-8000 to the new conical. Gently mix
- 5.Incubate at 4°C for at least 60 minutes
- 6.Centrifuge at 12,000 x g for 10 minutes. Carefully decant
- 7.Spin again briefly
- 8.Gently resuspend pellet in 1.6mL 1x TBS
- 9.Aliquot 800ul into 2 microfuge tubes. Proceed with both tubes
- 10.Spin sample for 1 minute to pellet remaining cells. Transfer supernatant to fresh tubes
- 11.Add 160ul of 2.5M NaCl/20% PEG-8000 solution to each
- 12.Let sit at room temperature for 5 minutes
- 13.Spin at 1300 x g for 10 minutes
- 14.Decant the supernatant
- 15.Spin briefly. Remove supernatant with pipet
- 16.Resuspend pellets in 300ul TE
- 17.Phenol extraction: add 300ul phenol. Vortex for 15 seconds
- a.Let sit for 15 minutes. Spin for 10 minutes
- 18.Add H2O so volume samples is about 300ul
- 19.Phenol/chloroform extraction*: add 300ul PCIA
- a.Vortex for 15 seconds. Spin for 10 minutes
- 20.Repeat Phenol/Chloroform extraction
- 21.Chloroform extraction*: add 300ul chloroform
- a.Vortex 15 seconds. Spin for 10 minutes
- 22.Add 30ul 2.5M NaAc (pH 4.8)
- 23.Add 2-2.5 volumes ethanol
- 24.Let precipitate for ~2 hours at -20°C
- 25.Spin for 1 minute
- 26.Decant supernatant
- 27.Resuspend in 25-50ul TE
- Performing steps at 4°C helps with separation
Protocol from: “Use of M13K07 Helper Phage for isolation of single-stranded phagemid DNA” by NEB
M13 Amplification
This protocol is to make more M13 phages.
Need
- Fresh plate of infectable cells (contain F’ episome)
Day 1
- 1.Grow liquid overnight culture of infectable cells
Day 2
- 1.Add 200ul overnight culture to 20mL LB in a 250mL flask
- 2.Add 1ul phage suspension
- 3.Incubate for 4-5 hours at 37°C, 250rpm
- 4.Centrifuge for 10 minutes at 4500 x g
- 5.Transfer supernatant to a new tube.
- 6.Repeat centrifugation on supernatant
- 7.Transfer top 16mL of supernatant to a new tube
- 8.Add 4mL of 2.5M NaCl/20% PEG-8000. Briefly mix
- 9.Precipitate phage for 1 hour or overnight at 4°C
- 10.Centrifuge for 15 minutes at 12000 x g. Decant supernatant
- a. Spin briefly. Remove residual supernatant with pipet
- 11.Resuspend pellet in 1mL 1x TBS. Transfer to 1.7mL tube
- 12.Spin briefly to remove cell debris
- 13.Transfer supernatant to a new tube
- 14.Add 200ul of 2.5M NaCl/20% PEG-8000
- 15.Incubate on ice for 15-60 minutes
- 16.Spin for 10 minutes at 12000-14000 rpm. Discard supernatant
- 17.Briefly spin. Remove supernatant with pipette
- 18.Resuspend pellet in 200uL TBS
- 19.For long term storage at -20C, add 200uL glycerol
Adapted from M13 Amplification protocol from NEB
Mini-prep
Notes before starting
- Optional: Add LyseBlue reagent to Buffer P1 at a ratio of 1 to 1000
- Add the provided RNase A solution to Buffer P1, mix, store bottle at 2-8°C
- Add ethanol (96-100%) to Buffer PE before use
- All centrifugation steps are carried out at 13,000 rpm (~17,900xg) in a conventional table-top microcentrifuge
Mini-prep
- 1.Centrifuge 1-6mL bacterial overnight culture at >8000 rpm (6800xg) for 3 minutes at room temperature (15-25C)
- 2.Resuspend pellet in 250ul Buffer P1 and transfer to microcentrifuge tube
- 3.Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear.
- a.DO NOT allow lysis reaction to proceed for more than 5 minutes
- b.If using LyseBlue reagent, the solution will turn blue
- 4.Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
- a.If using LyseBlue reagent, the solution will turn colorless
- 5.Centrifuge for 10 minutes
- 6.Apply supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60 s and discard the flow-through
- 7.Recommended: Wash the QIAprep spin column by adding 500 ul Buffer PB. Centrifuge for 30-60 s and discard the flow-through
- a.Only required when using endA+ strains or other bacterial strains with high nuclease activity or carbohydrate content
- 8.Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 30-60 s and discard the flow-through
- 9.Centrifuge for 1 minutes to remove residual wash buffer
- 10.Place the QIAprep column in a clean 1.5mL microcentrifuge tube. To elute DNA, add 30ul Buffer EB. Let stand for 1 min, and centrifuge for 1 minute
- a.Can elute DNA in 50ul but this will decrease DNA concentration
- b.To increase yield, let sit for up to 4 minutes
Protocol and Reagent recipes from QIAGEN unless stated otherwise
Bacterial Infection
Need
- Fresh plate of cells that contain the F’ episome
Day 1
- 1.Pick colony from plate and grow overnight
Day 2
- 1.Dilute overnight to OD600 0.1
- 2.Incubate until OD600 ~ 0.5*
- 3.Add phage to a final concentration of 1 x108 phage/mL
- 4.Incubate for 30 minutes to infect
- 5.Plate samples under selection
- a.We recommend trying a range of dilutions
- Note: All incubations are done at 37°C and shaking at 225rpm
- Note: If multiple samples are to be done in parallel
- If using the same cells, we suggest growing one large batch of cells. Once the OD has reached ~0.5, divide into 50mL samples then add phage.
- If using different cells, grow each sample to ~0.5 then store on ice. Once all samples have reached ~0.5, incubate on ice for another 30 minutes. Warm for 20-30 minutes at 37C, 250 rpm. Measure OD again to check that samples are comparable (yes, they will have grown some). Add phage
Protocol from “Eliminating Helper phage from Phage Display
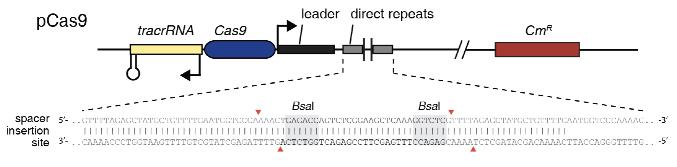
Cloning a new spacer into pCas9
Design the new spacer
- 1. Find a PAM site (5’- NGG -3’). This will be on the 3’ end of the targeting sequence
- 2. The 30bps upstream of NGG motif (on the same strand) will be the spacer
- 3. Oligo I (fwd. primer): To the spacer sequence, add AAAC to the 5’ end and G to the 3’ end
- a. NOTE: These nucleotides are for ligation purposes and are NOT used in targeting. Therefore, they do not have to be present in the target sequence and do not count towards the 30bps
- 4. Oligo II (rvs. primer): Find the reverse complement of the spacer sequence. Add CAAAA to the 5’ end.
Phosphorylation
- 1.Mix the following reactants
| Reactant | Amount |
|---|---|
| Oligo I (100uM) | 1 ul |
| Oligo II (100uM) | 1 ul |
| 10x T4 Ligase Buffer | 5 ul |
| T4 PNK | 1 ul |
| H2O | 42 ul |
- 2.Incubate for 60 minutes at 37C then 20 minutes at 65C
Anneal Oligos
- 1. Add 2.5 ul of 1M NaCl to the phosphorylation product
- 2. Incubate for 5 minutes at 95C then let slowly cool to room temperature.
- a. It is recommended to use a thermocycler that either cools at a gradient or steps down in temperature over the course of ~2 hours.
- b. Alternatively, incubate the sample at 95C for 5 minutes then remove the heat block from the heater and let it cool naturally for 2 hours
- 3. Dilute the annealed oligos 1:10
Vector digest with BsaI
- 1. Mix the following reactants
| Reactant | Amount |
|---|---|
| Cutsmart Buffer | 5 ul |
| BsaI enzyme | 1 ul |
| DNA | 1-2ug |
| H2O | Bring volume to 50ul |
- 2. Incubate for 90 minutes at 37C then 20 minutes at 65C
- 3. Gel extract digested pCas9
Ligation
- 1. Mix the following reactants
| Reactant | Amount |
|---|---|
| Gel extracted vector | 1 ul |
| Diluted annealed oligos | 2 ul |
| 10x T4 Ligase Buffer | 2 ul |
| T4 Ligase | 1 ul |
| H2O | 14 ul |
- 2. Incubate overnight (8-12 hours) at 16C
Transform
- 1. Transform into DH5alpha
Protocol from Marraffini Lab
 "
"