Team:CU-Boulder/Notebook/CC9Phage Team
From 2014.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
{{UCB-NavBar}} | {{UCB-NavBar}} | ||
__FORCETOC__ | __FORCETOC__ | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
'''Note:''' | '''Note:''' | ||
'''Litmus28ibb is a biobricked version of the Litmus28i (from NEB) vector which contains the M13ori (packaging signal) which allows for uptake into M13 or Fd phage. While making this part biobrick compatible, we added J04450 as the insert.''' | '''Litmus28ibb is a biobricked version of the Litmus28i (from NEB) vector which contains the M13ori (packaging signal) which allows for uptake into M13 or Fd phage. While making this part biobrick compatible, we added J04450 as the insert.''' | ||
'''pCas9 (BBa_K1218011) contains and tracrRNA and Cas9 gene under native promoters, and a minimal CRISPR arrays. The guide RNA (referred here as gRNA) is inserted into the CRISPR array.''' | '''pCas9 (BBa_K1218011) contains and tracrRNA and Cas9 gene under native promoters, and a minimal CRISPR arrays. The guide RNA (referred here as gRNA) is inserted into the CRISPR array.''' | ||
| + | |||
| + | ==Week 1== | ||
| + | |||
'''7/21''' | '''7/21''' | ||
*To ligate pCas9 to Litmus28ibb | *To ligate pCas9 to Litmus28ibb | ||
| Line 45: | Line 52: | ||
**Ligation (10hrs at 16C, 10min at 80C) | **Ligation (10hrs at 16C, 10min at 80C) | ||
***Litmus28ibb + pCas9 + J04450 | ***Litmus28ibb + pCas9 + J04450 | ||
| + | |||
| + | ==Week 2== | ||
'''7/27''' | '''7/27''' | ||
| Line 107: | Line 116: | ||
**colonies from 8/2 transformation | **colonies from 8/2 transformation | ||
**restreak of LCR from freeze down | **restreak of LCR from freeze down | ||
| + | |||
| + | ==Week 3== | ||
'''8/4''' | '''8/4''' | ||
| Line 168: | Line 179: | ||
***pSB1C3-pCas9(unmod.) | ***pSB1C3-pCas9(unmod.) | ||
**Transform and plate onto low Chlor 934 ug/mL) | **Transform and plate onto low Chlor 934 ug/mL) | ||
| + | |||
| + | ==Week 4== | ||
'''8/10''' | '''8/10''' | ||
| Line 353: | Line 366: | ||
*PCR on LCR (Litmus28ibb with pCas9 and RFP to maybe get a good product to religate | *PCR on LCR (Litmus28ibb with pCas9 and RFP to maybe get a good product to religate | ||
**Didn’t work | **Didn’t work | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
Revision as of 04:36, 13 October 2014
Note: Litmus28ibb is a biobricked version of the Litmus28i (from NEB) vector which contains the M13ori (packaging signal) which allows for uptake into M13 or Fd phage. While making this part biobrick compatible, we added J04450 as the insert. pCas9 (BBa_K1218011) contains and tracrRNA and Cas9 gene under native promoters, and a minimal CRISPR arrays. The guide RNA (referred here as gRNA) is inserted into the CRISPR array.
Contents |
Week 1
7/21
- To ligate pCas9 to Litmus28ibb
- Digestion
- Litmus28ibb-J04450 with E+P
- pCas9 with E+P
- Digestion
- 1. pCas9
- 2. Litmus28ibb
- Overnight ligations (10hrs at 37C, 10min at 80C)
7/22
- Transformed ligation from 7/21 (Litmus28ibb + pCas9) into ER2738 cells
- Plated on Amp + Tet
- Later in day, pricked colonies from morning transformation (Litmus28ibb and pCas9 in ER)
7/23
- Miniprepped O/N from 7/22 of Litmus28ibb-pCas9
- Digested
- None were the correct size. Smaller bands looked like pSB1C3. Plated cells onto Chlor and there was significant growth. pSB1C3 from the pCas9?
- Digested
7/24
- To put pCas9 onto Litmus28i
- Digested
- Litmus28ibb with EcoRI+PstI
- pSB1C3-pCas9 with EcoRI+SpeI
- PSB3C5-J04450 with XbaI + PstI
- Digested
- 1. Litmus28ibb
- 2. pCas9
- 3. J04450
- Overnight ligation (10hr @ 16C)
7/25
- Transformed ligation from 7/24 (Litmus28ibb+pCas9) into 5alpha cells
7/26
- No growth from 7/25 transformation
- Tranform
- Litmus28ibb + pCas9 + J04450
- Retry making Litmus28i with pCas9
- Ligation (10hrs at 16C, 10min at 80C)
- Litmus28ibb + pCas9 + J04450
- Ligation (10hrs at 16C, 10min at 80C)
Week 2
7/27
- Transformation results
- No growth for no DNA controls **4 colonies for Litmus28bb-JO4450-Cas9 *Picked all 4 colonies for O/Ns
7/28
- 3 new red colonies on Litmus28bb-Cas9-J04450 plate
- Set up cultures
- O/Ns grew but did not bother
- Transformations
- Litmus28bb-J04450-Cas9
- No DNA control
7/29
- Litmus28bb-Cas9-J04450 O/Ns
- Incubator shut off during night. Samplers were at RT by morning
- Add 1mL to fresh 4mL LB and Amp
- Mini-prep DNA
- Digest with EcoRI and PstI to check insert for correct size
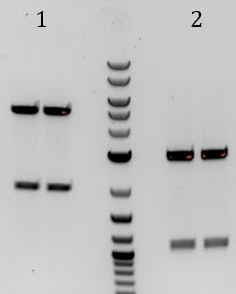
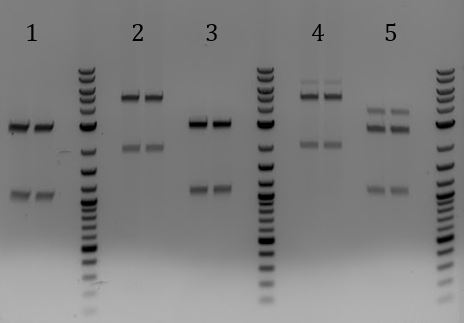
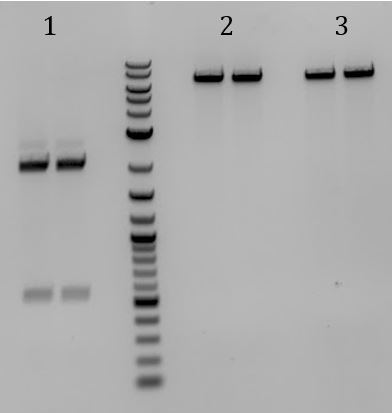
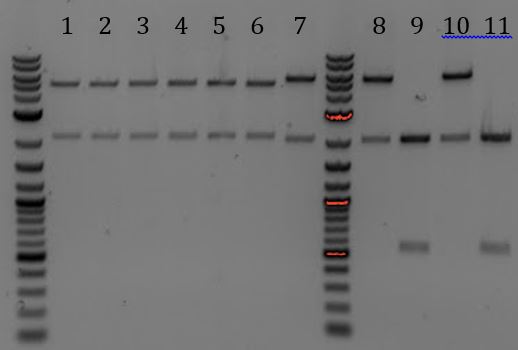
- Lane 2 contains the expected 3000bp band for the Litmus28ibb backbone and the ~7000bp band for the pCas9-J04450 insert. Unfortunately, this sample also contains a 4000bp band.
- Plated the above sample
7/30
- Digested Litmus28ibb-Cas9-RFP with EcoRI-HF
- Extracted large band ~9000
- Re-ligated piece
- Sent samples for sequencing
- Litmus28bb with Cas9 and RFP (LCR) 8/1
- Mini-prepped O/Ns of
- Litmus28ibb-Cas9-J04450 (LCR)
- Digested with EcoRI-HF and PstI-HF => all samples had band ~3700
- Litmus28ibb-Cas9-J04450 (LCR)
- Digestions of Litmus and Cas9 to retry ligation
- 1. Litmus28ibb (E+P)
- 2. pCas9(E+P)
- 3. Litmus28ibb (E+P)
- 4. pCas9(E+S)
- 5. pSB3C5 (X+P)
- Gel extractions
- Ligations
- Litmus28ibb (E+P) to pCas9 (E+P)
- Litmus28ibb(E+P) to pCas9(E+S) and J04450(E+P)
8/2
- Transformation into BW cells (5/30)
- Hundreds of colonies for LCR Miniprep
- Digestion to get a better idea of what this DNA is
8/3
- Isolate the LCR from the extra band in the gel
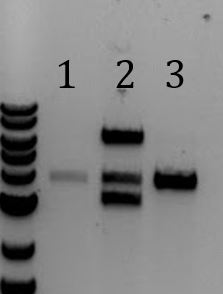
- Run 8/2 digestions on gel
- 1. LCR***2. LCR**
- Gel extracted
- LCR digested with PstI-HF (top band
- Re-ligate extracted LCR with no insert (should re-ligate to itself)
- Transformed ligation into ER2738
- Made O/Ns
- colonies from 8/2 transformation
- restreak of LCR from freeze down
Week 3
8/4
- Check overnights from 8/3 (LCR = Litmus28ibb+pCas9+RFP) for correct inserts
- Mini-prep samples
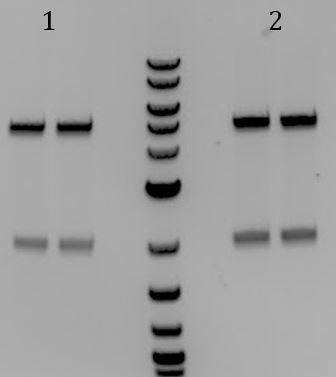
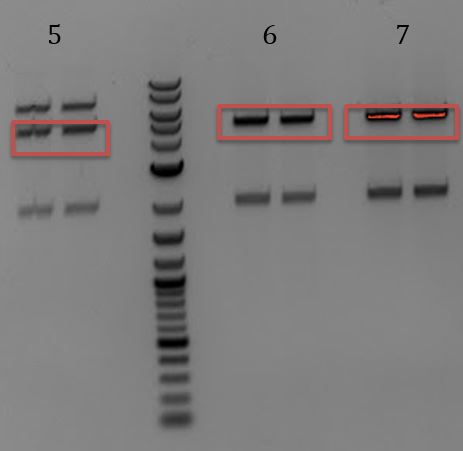
- Digest with EcoRI and PstI
- All had sizes ~3700bp
- No correct inserts
- Attempt to ligate Litmus28ibb and pCas9(gRNA3)
- Digestions
- Litmus28ibb-RFP E+P extracted Lit back bone
- pCas9 gRNA3 E+P extracted pCas9 insert
- Digestions
- 1. Litmus28ibb
- 2. pCas9(gRNA3)
- 3. pCas9(unmod.)
- Ligations (10 hrs at 16C, 10 min at 80C)
- Litmus28ibb + pCas9(gRNA3)
- Ligations (10 hrs at 16C, 10 min at 80C)
8/5
- Transformation into ER2738 (7/18), 5ul of ligation products
- 1. Litmus28ibb-Cas9gRNA3
8/6
- Another attempt at ligating pCas9 and Litmus28ibb
- Digest unmodified pCas9 with EcoRI-HF and PstI-HF
- Digest pCas9 containing pRNA1 with EcoRI-HF and PstI-HF
- 1. pCas9(unmod.)
- 2. pCas9(gRNA1)
- Gel extract pieces
- Ligate separately to Litmus28ibb
- Results of 8/5 transformation
- 6 colonies for Litmus28ibb+pCas9(gRNA3) on Tet+reduced Amp
- Picked colonies for overnights
- No growth for no DNA control
- 6 colonies for Litmus28ibb+pCas9(gRNA3) on Tet+reduced Amp
- Transformation of ligations from earlier today
- Litmus28ibb + pSB1C3-pCas9(unmod.)
- Litmus28ibb + pSB1C3-pCas9(gRNA1)
- Mini-prep Litmus28ibb+pCas9(gRNA3) O/N samples from earlier in day
- Digested with EcoRI-HF and PstI-HF
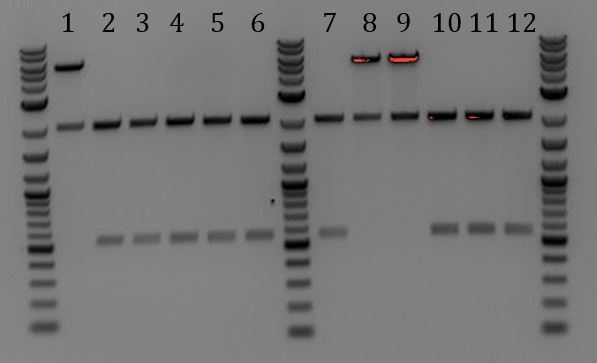
- All samples had bands of 3000 and 2000bp. None were correct
- Digested with EcoRI-HF and PstI-HF
8/8
- M13ori into SB Cas9
- Digestions
- unmodified pCas9 with EcoRI-HF and XbaI
- M13ori with EcoRI-HF and SpeI-HF
- pCas9(gRNA1) with EcoRI-HF and XbaI
- pCas9(gRNA3) with EcoRI-HF and XbaI
- Digestions
8/9
- To clone M13ori onto pSB1C3-pCas9
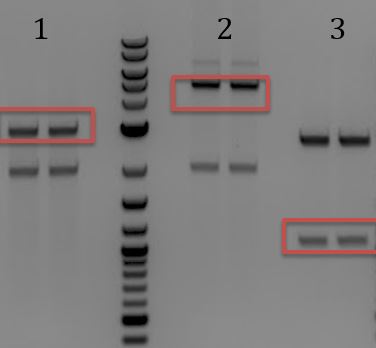
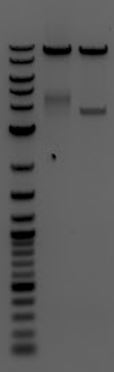
- Run 8/8 digestion on a gel
- 1. M13ori
- 2. pCas9(gRNA3)
- 3. pCas9(gRNA1)
- Gel extracted
- Ligation Ligated M13ori to….
- pSB1C3-pCas9(gRNA1)
- pSB1C3-pCas9(gRNA3)
- pSB1C3-pCas9(unmod.)
- Transform and plate onto low Chlor 934 ug/mL)
Week 4
8/10
- Digested ligation products from 8/6 and 8/4. Saw many large bands that may have been undigested
- O/Ns of 8/9 transformation
- 1 colony for M13ori-pCas9(gRNA3) (only 1 colony grew)
- 11 colonies for M13ori-pCas9(gRNA1)
- No colonies grew for M13ori-pCas9(unmodified)
8/11
- Test 8/10 O/Ns for correct inserts
- Digest 8/10 O/Ns with EcoRI-HF and PstI-HF
- 1. M13ori-pCas9(gRNA3)
- 2-12. M13ori-pCas9(gRNA1)
- Lanes 1, 8, and 9 are about the right size. Sent these samples for sequencing
- To ligate M13ori to pSB1C3-pCas9(unmod.)
- Digest
- pSB1C3-pCas9(unmod.) E+P
- M13 digested 8/8
- Ligation: See L5 below
- Digest
- To ligate pCas9 and M13ori to pSB1A3
- Digest
- pSB1A3 ***M13ori ***Unmodified pCas9 ***pCas9(gRNA1)-M13ori ***pCas9(gRNA3)-M13ori **Ligation: See L2-4 below
- Digest
- Another try to put pCas9 onto Litmus28ibb
- Digested
- Unmodified Litmus28i ***pCas9(unmod.) ***pCas9(gRNA1)-M13ori E+P
- pCas9(gRNA3)-M13ori E+P
- Ligation: See L1 and L6 below
- Digested
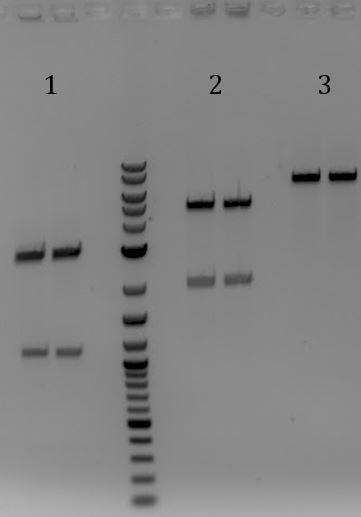
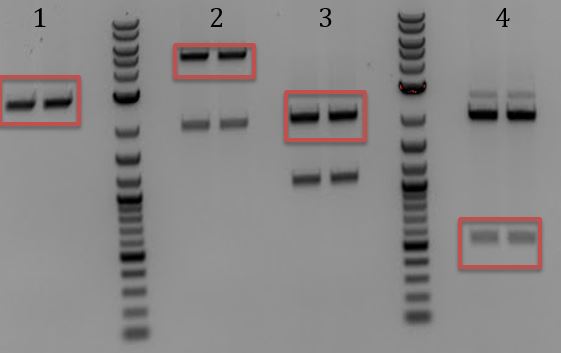
- Gel for all
- 1. Unmodified Litmus28i
- 2. pSB1C3-pCas9(unmodified)
- 3. pSB1A3
- 4. pSB1C3-M13ori
- 5. pSB1C3-pCas9(unmod.)
- 6. pSB1C3-M13ori-pCas9(gRNA1)
- 7. pSB1C3-M13ori-pCas9(gRNA3)
- Ligations for both projects
| # | Backbone | Insert 1 | Insert 2 |
|---|---|---|---|
| 1 | Unmod. Litmus28i (E+P) | pCas9(unmod.) (E+P) | N/A |
| 2 | pSB1A3 (E+P) | pCas9(unmod.) (X+P) | M13ori (E+S) |
| 3 | pSB1A3 (E+P) | pCas9(gRNA1)-M13ori (E+P) | N/A |
| 4 | pSB1A3 (E+P) | pCas9(gRNA3)-M13ori (E+P) | N/A |
| 5 | pSB1C3-pCas9(unmod.) (E+X) | M13ori (E+S) | N/A |
| 6 | Litmus28ibb (E+P) | pCas9(gRNA3) (E+P) | N/A |
| 7 | Litmus28ibb (E+P) | pCas9(gRNA1) (E+P) | N/A |
| 8 | Litmus28ibb (E+P) | pCas9(unmod.) (E+P) | N/A |
8/12
- Transform ligation from 8/11 into ER2738 cells
- Make phage using M13g6A1 as Helper Phage
- used pCas9(gRNA1)-M13ori or pCas9(gRNA3)-M13ori as phagemid
- Freeze downs
| Top | Side |
|---|---|
| M. gRNA3 Cas9 (8/12) | M13ori Cas9 gRNA3 pSB1C3 |
| M. gRNA1 Cas9 (8/12) | M13ori(8) Cas9 gRNA1 pSB1C3 |
| M. gRNA1 Cas9 (8/12) | M13ori (9) Cas9 gRNA1 pSB1C3 |
8/13
- Finish phage isolation
| Helper phage | Phage | A269 | A320 | genome size | [ ] |
|---|---|---|---|---|---|
| Fd-CAT | M. gRNA1 | 0.353 | 0.025 | 7687 | 2.56 x10^12 |
| Fd-CAT | M. gRNA3 | 0.554 | 0.037 | 7687 | 4.40 x10^12 |
- Infection
- Infecting ER2738 and BWF’
- Phage
- pSB1C3-pCas9(gRNA1)
- pSB1C3-pCas9(gRNA3)
- Transformation Results from 8/12
- Growth for the following… (made O/Ns)
- pSB1A3-M13ori-pCas9(gRNA3)
- pSB1C3-M13ori-pCas9(unmod)-M13ori
- Litmus28ibb-pCas9(gRNA3)
- Litmus28ibb-pCas9(gRNA1)
- Litmus28ibb-pCas9(unmod.)
- Hundreds of colonies for pCas9(unmod.) on Unmod. Litmus28i. Didn’t trust result so didn’t pick coloneis. EcoRI and PstI sites touch so most likely, Litmus28i was only cut once
- Growth for the following… (made O/Ns)
8/15
- Check O/Ns from 8/14 for correct inserts
- Mini-prepped samples
- Digested with E and P
- Results
- No bands of the right size for
- Litmus28ibb-pCas9(gRNA3)
- Litmus28ibb-pCas9(gRNA1)
- Litmus28ibb-pCas9(unmod.)
- Expected band sizes for… (see below)
- pCas9(gRNA3)-M13ori on pSB1A3
- pCas9(unmod)-M13ori on pSB1C3
- Transform into BWF’ to test for functionality of Cas9 when associated with m13ori, independent of infectivity
- M13ori-pCas9(gRNA1)
- M13ori-pCas9(gRNA3)
8/16
- PCR on LCR (Litmus28ibb with pCas9 and RFP to maybe get a good product to religate
- Didn’t work
 "
"