Galectin-3 - An Alternative Sensing Molecule
We are committed to constantly improve our detection methods. Therefore, we already thought ahead and came up with an alternative approach for the detection of pathogens. The current method uses the quorum sensing system pathogens and is thus limited to bacteria that secrete autoinducers. Our alternative detection system involves biomolecules tagged with a reporter that bind to the surface of the cell and reveal its presence.
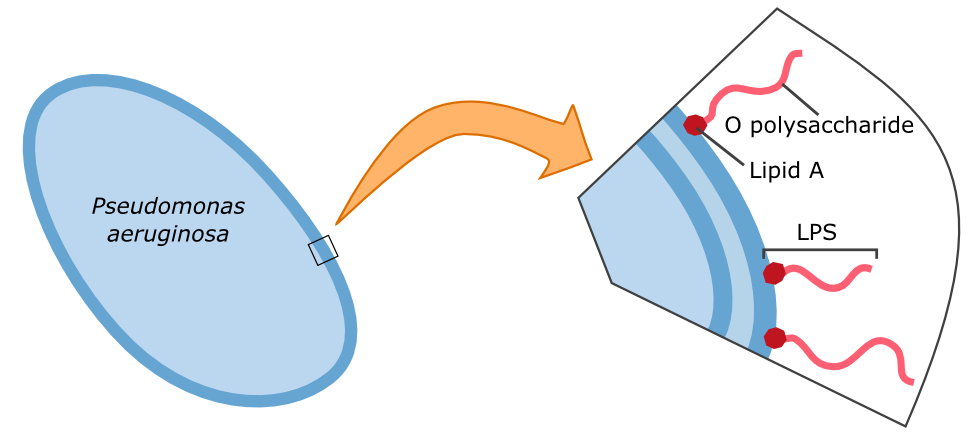
The specific binding of galectin-3 enables the construction of such a detection system. Parts of the lipopolysaccharide structure (LPS) of Pseudomonas aeruginosa can be bound by galectin-3. A fusion protein of galectin-3 and a reporter protein, such as a fluorescent protein, can be built and applied in the detection of Pseudomonas aeruginosa.
In our approach, a galectin-3-YFP fusion protein is built and expressed in E. coli. A his-tag and a snap-tag for purification are included. The fusion protein can then be incorporated into a cell-free biosensor system. Such biosensors have many advantages over systems that use living cells; storage, for example, is much easier. From a biosafety and social acceptance perspective, it is also advantageous if the sensor system does not contain live genetically modified organisms.
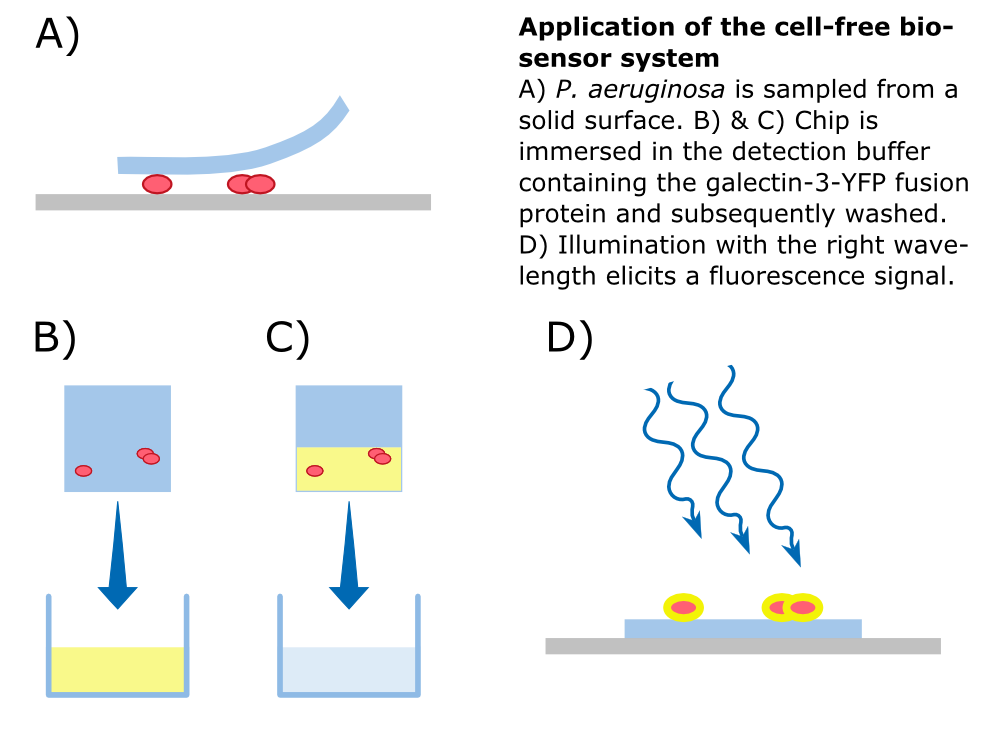
To detect P. aeruginosa cells, an agar chip could be used to sample a solid surface. However, other materials but agar can be considered to collect the pathogens. The cell stick to the sampling chip which is then immersed in a detection buffer containing the galectin-3-YFP fusion protein. Excess protein is removed during washing in a suitable buffer. The galectin-3 remains bound to the pathogen and illumination with 514 nm, the excitation frequency of YFP, in a modified version of our measurement device reveals the location of the cells. The picture taken by the measurement device can then be analyzed by our software Measurarty.
|
 "
"