Team:Aachen/Project/FRET Reporter
From 2014.igem.org
(→A Fluorescence Answer Faster Than Expression) |
m (→Comparing the kinetic of the double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 with standard GFP expression) |
||
| (168 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
{{Team:Aachen/Header}} | {{Team:Aachen/Header}} | ||
| - | =The REACh Construct= | + | =The modified REACh Construct= |
| - | On this page, we present our biosensor on | + | On this page, we present our biosensor on a molecular level. Here you can explore the different parts of our genetic device: |
<html> | <html> | ||
<center> | <center> | ||
| - | <ul class="menusmall-grid"> | + | <ul class="menusmall-grid" style="width:inherit;"> |
<!-- Overview --> | <!-- Overview --> | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#fluorescence" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#fluorescence" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">A Faster Answer</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">A Faster Answer</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/0/0b/Aachen_14-10-13_GFP_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/0/0b/Aachen_14-10-13_GFP_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
| Line 23: | Line 23: | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#fret" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#fret" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">The FRET System</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">The FRET System</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/5/54/Aachen_14-10-13_FRET_Arrows_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/5/54/Aachen_14-10-13_FRET_Arrows_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
| Line 31: | Line 31: | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#darkquencher" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#darkquencher" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">REACh Quenchers</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">REACh Quenchers</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/65/Aachen_14-10-13_REACh_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/65/Aachen_14-10-13_REACh_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
| Line 39: | Line 39: | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#gfp-reach" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#gfp-reach" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">The Fusion Protein</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">The Fusion Protein</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/0/02/Aachen_14-10-13_Fusion_Protein_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/0/02/Aachen_14-10-13_Fusion_Protein_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
</a> | </a> | ||
</li> | </li> | ||
| + | </ul> | ||
| + | </center> | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <ul class="menusmall-grid" style="width:516px;"> | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#tevprotease" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#tevprotease" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">TEV Protease</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">TEV Protease</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/d/dc/Aachen_14-10-13_TEV_Protease_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/d/dc/Aachen_14-10-13_TEV_Protease_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
| Line 55: | Line 62: | ||
<li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
<a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#reachachievements" style="color:black"> | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#reachachievements" style="color:black"> | ||
| - | <div class="menusmall-item menusmall-info" ><div class="menukachel">Achieve-<br/>ments</div></div> | + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:32%; line-height:1.5em;">Achieve-<br/>ments</div></div> |
<div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/e/ef/Aachen_14-10-15_Medal_Cellocks_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/e/ef/Aachen_14-10-15_Medal_Cellocks_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
| + | </div> | ||
| + | </a> | ||
| + | </li> | ||
| + | |||
| + | <li style="width: 156px;margin-left: 8px;margin-right: 8px;"> | ||
| + | <a class="menulink" href="https://2014.igem.org/Team:Aachen/Project/FRET_Reporter#reachoutlook" style="color:black"> | ||
| + | <div class="menusmall-item menusmall-info" ><div class="menukachel" style="top:40%;">Outlook</div></div> | ||
| + | <div class="menusmall-item menusmall-img" style="background: url(https://static.igem.org/mediawiki/2014/6/67/Aachen_14-10-16_Outlook_Cellocks_iNB.png); norepeat scroll 0% 0% transparent; background-size:100%"> | ||
</div> | </div> | ||
</a> | </a> | ||
| Line 70: | Line 85: | ||
[[File:Aachen_14-10-13_GFP_iNB.png|150px|right]] | [[File:Aachen_14-10-13_GFP_iNB.png|150px|right]] | ||
| - | = A Fluorescence Answer Faster | + | = A Fluorescence Answer Faster than Expression = |
<span class="anchor" id="fluorescence"></span> | <span class="anchor" id="fluorescence"></span> | ||
| - | + | ||
| + | Biosensor systems are usually based on a reporter gene under the control of a promoter directly induced by the biomolecule of interest. In the case of our 2D biosensor for ''Pseudomonas aeruginosa'', the expression of our fluorescing reporter protein is directly induced by the activity of the bacterium's quorum sensing molecules. However, transcription, translation, folding and post-translational modifications take their time. Since our goal is to detect the pathogen as fast as possible, we aimed to develop a system that provides a fluorescent answer faster than just expressing the fluorescent protein GFP. | ||
<center> | <center> | ||
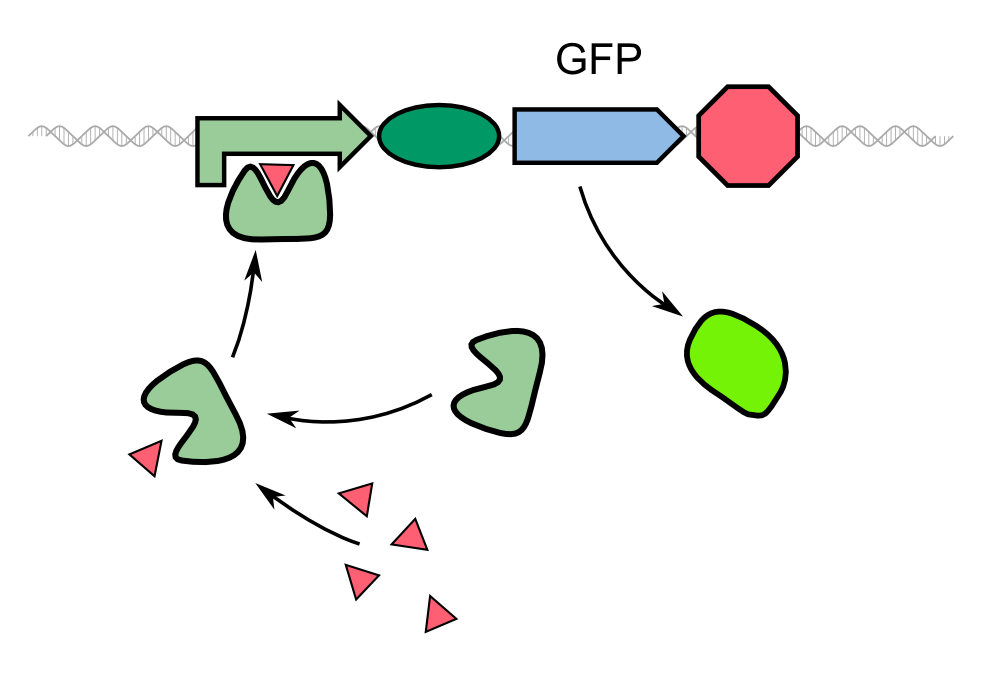
| - | {{Team:Aachen/Figure|Aachen_Traditional_biosensor.png|align=center|title= | + | {{Team:Aachen/Figure|Aachen_Traditional_biosensor.png|align=center|title=Schematic model of a traditional biosensor|subtitle=In this model, the expression of GFP is directly controlled by a promoter whose activator binds to a molecule secreted by the pathogen.|width=500px}} |
</center> | </center> | ||
| - | + | As an alternative to the traditional approach, we '''constitutively express our reporter gene in a quenched form'''. By fusing GFP to REACh, a mutated variant of EYFP, its original fluorescence is thoroughly suppressed. Thereby, a huge stock of temporary inactivated fluorescence proteins is already available prior to first contact with the pathogen. As a conseqence to the uptake of homoserine lactones of ''P. aeruginosa'' and binding to the LasI promoter in front of the protease gene, these '''autoinducers activate the expression of the TEV protease'''. This way, our biosensor provides a fast increase in fluorescence intensity by cleavage of GFP-REACh-constructs. | |
<center> | <center> | ||
| - | {{Team:Aachen/Figure|Aachen_REACh_approach.png|align=center|title= | + | {{Team:Aachen/Figure|Aachen_REACh_approach.png|align=center|title=Schematic model of our novel biosensor|subtitle=Expression of the TEV protease is induced by HSL. The protease cleaves the GFP-REACh fusion protein to elicit a fluorescence response.|width=900px}} |
</center> | </center> | ||
| - | + | '''Advantages of the novel biosensor:''' | |
| - | + | ||
| - | * | + | * When ''P. aeruginosa'' is detected by our cells, the reporter protein has already been expressed and only waits to be activated. The cleavage reaction catalyzed by the TEV protease is a faster process than expression and correct folding of GFP. Therefore, our sensor cells provide an exceptionally '''early response'''. |
| - | + | * While a certain concentration of homoserine lactone will produce the same number of gene read-outs and the TEV protease is about the same size as GFP (~27 kDa), one TEV protease can cleave many GFP-REACh constructs per time than fluorescent protein molecules could be expressed and go through fluorescence maturation. Through this additional reaction, we introduce an '''amplification step''' into our system. Hypothetically, the TEV protease will enable the production of '''a much stronger signal''' in a given time interval. | |
| - | + | ||
| - | + | In parallel, we worked on a [https://2014.igem.org/Team:Aachen/Project/Model model] of the respective biosensor system, in order to confirm our hypothesis of a faster and stronger fluorescence response than a conventional biosensor would povide. | |
| - | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 104: | Line 118: | ||
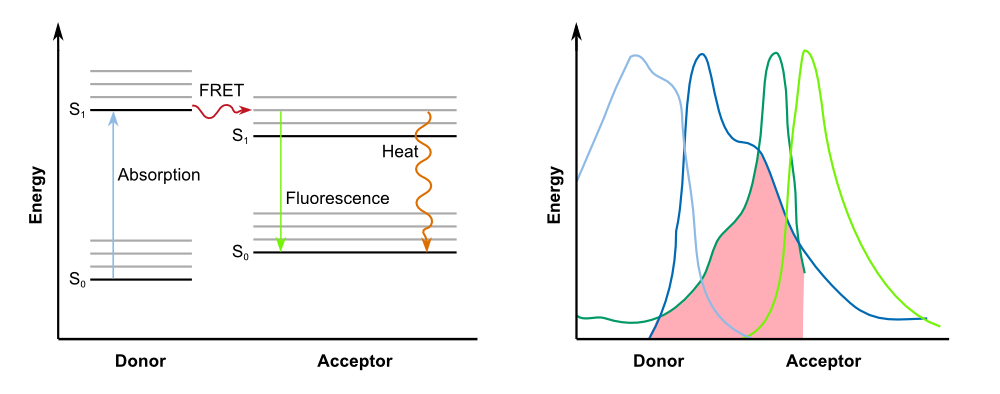
Förster resonance energy transfer (FRET), sometimes also called fluorescence resonance energy transfer, is a physical process of energy transfer. In FRET, the energy of a donor chromophore, whose electrons are in an excited state, is passed to a second chromophore, the acceptor. The '''energy is transferred without radiation''' and is therefore not exchanged via emission and absorption of photons. The acceptor then releases the energy received from the donor, for example, as light of a longer wavelength. | Förster resonance energy transfer (FRET), sometimes also called fluorescence resonance energy transfer, is a physical process of energy transfer. In FRET, the energy of a donor chromophore, whose electrons are in an excited state, is passed to a second chromophore, the acceptor. The '''energy is transferred without radiation''' and is therefore not exchanged via emission and absorption of photons. The acceptor then releases the energy received from the donor, for example, as light of a longer wavelength. | ||
| - | In biochemistry and cell biology | + | In biochemistry and cell biology fluorescent dyes, which interact via FRET, are applied as "optical nano metering rules", because the intensity of the transfer is dependent on the spacing between donor and acceptor. FRET can be observed over distances of up to 10 nm. This way, protein-protein interactions and conformational changes of a variety of tagged biomolecules can be observed. For an efficient FRET to occur, there must be a substantial overlap between the donor fluorescence emission spectrum and the acceptor fluorescence excitation (or absorption) spectrum ([http://www.nature.com/nprot/journal/v8/n2/full/nprot.2012.147.html Broussard et al., 2013]), as described in the figure below. |
{{Team:Aachen/Figure|Aachen_14-10-07_Jablonski_Diagram_and_Absorption_Spectra_iNB.png|align=center|title=FRET betweeen donor and acceptor|subtitle=On the left: Jablonski diagram showing the transfer of energy between donor and acceptor; on the right: For successful FRET, the emission spectrum of of the donor has to overlap with the absorption spectrum of the acceptor.|width=900px}} | {{Team:Aachen/Figure|Aachen_14-10-07_Jablonski_Diagram_and_Absorption_Spectra_iNB.png|align=center|title=FRET betweeen donor and acceptor|subtitle=On the left: Jablonski diagram showing the transfer of energy between donor and acceptor; on the right: For successful FRET, the emission spectrum of of the donor has to overlap with the absorption spectrum of the acceptor.|width=900px}} | ||
| - | However, '''fluorescence is not an essential requirement for FRET'''. This type of energy transfer can also be observed between donors that are capable of other forms of radiation, such as phosphorescence, bioluminescence or chemiluminescence | + | However, '''fluorescence is not an essential requirement for FRET'''. This type of energy transfer can also be observed between donors that are capable of other forms of radiation, such as phosphorescence, bioluminescence or chemiluminescence. Acceptor chromophores do not necessarily emit the energy in form of light, and can lead to quenching instead. Thus, this kind of acceptors are also referred to as '''dark quenchers'''. In our project, we use a FRET system with a dark quencher, namely our '''REACh construct'''. |
| + | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 121: | Line 136: | ||
</center> | </center> | ||
| - | In 2006, [http://www.pnas.org/content/103/11/4089.full Ganesan et al.] were the first to present a previously undescribed FRET acceptor, a non-fluorescent yellow fluorescent protein (YFP) mutant called '''REACh ( | + | In 2006, [http://www.pnas.org/content/103/11/4089.full Ganesan et al.] were the first to present a previously undescribed FRET acceptor, a non-fluorescent yellow fluorescent protein (YFP) mutant called '''REACh (Resonance Energy-Accepting Chromoprotein)'''. YFP can be used as a FRET acceptor in combination with GFP as the donor in FRET microscopy and miscellaneous assays in molecular biology. The ideal FRET couple should possess a large spectral overlap between donor emission and acceptor absorption - as illustrated in previous section - but have separated emission spectra to allow their selective imaging. |
| + | |||
| + | To optimize the spectral overlap of this FRET pair, the group obtained '''a genetically modified YFP acceptor'''. Mutations of amino acid residues that stabilize the excited state of the chromophore in enhanced YFP (EYFP) resulted in a non-fluorescent chromoprotein. Two mutations, H148V and Y145W, reduced the fluorescence emission by 82 and 98%, respectively. Ganesan et al. chose the Y145W mutant and the Y145W/H148V double mutant as FRET acceptors, and named them REACh1 and REACh2, respectively. '''Both REACh1 and REACh2 act as dark quenchers of GFP'''. | ||
| - | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 132: | Line 148: | ||
<span class="anchor" id="gfp-reach"></span> | <span class="anchor" id="gfp-reach"></span> | ||
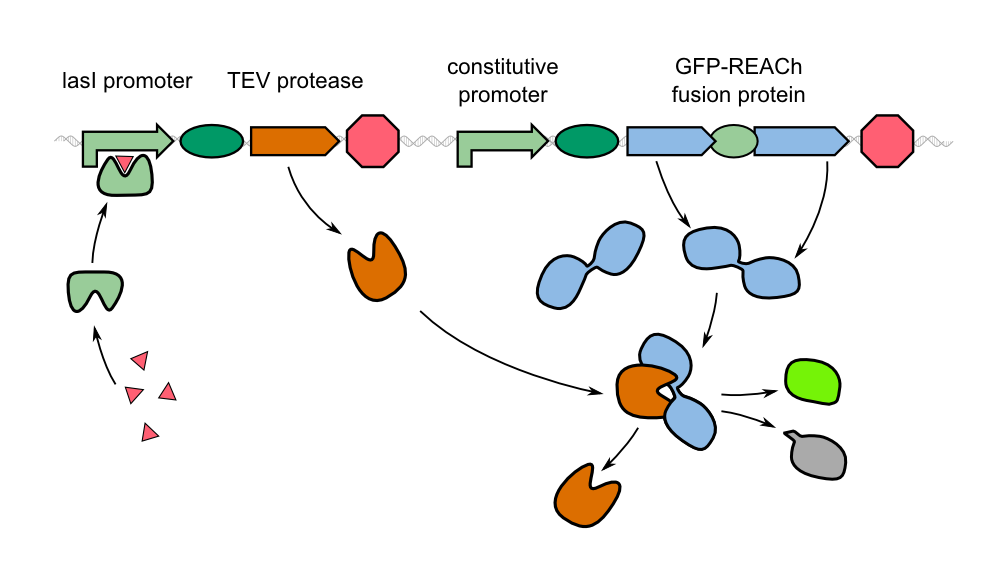
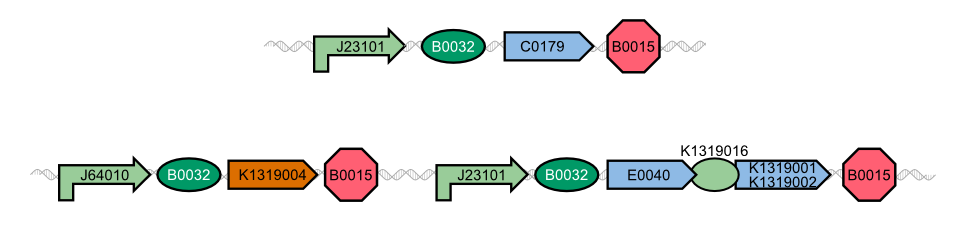
| - | In our project, we reproduced the REACh1 and REACh2 proteins by subjecting an RFC-25 compatible version of the BioBrick [http://parts.igem.org/Part:BBa_E0030 E0030] (EYFP) to a '''QuikChange mutation''', creating the BioBricks K1319001 and K1319002 | + | In our project, we reproduced the REACh1 and REACh2 proteins by subjecting an RFC-25 compatible version of the BioBrick [http://parts.igem.org/Part:BBa_E0030 E0030] (EYFP) to a '''QuikChange mutation''', creating the BioBricks [http://parts.igem.org/Part:BBa_K1319001 K1319001] and [http://parts.igem.org/Part:BBa_K1319002 K1319002]. Subsequently, we fused each REACh protein with '''GFP (mut3b)''' which is available as BioBrick [http://parts.igem.org/Part:BBa_E0040 E0040]. The protein complex was linked via a '''protease cleavage site''' ([http://parts.igem.org/Part:BBa_K1319016 K1319016]). As constitutive promoter we use [http://parts.igem.org/Part:BBa_J23101 J232101]. When GFP is connected to either REACh quencher, GFP will absorb light but the energy will be transferred to REACh via FRET and is then emitted in the form of heat; the fluorescence is quenched. Our cells also constitutively express the '''[http://parts.igem.org/Part:BBa_C0179 LasR] activator'''. Together with the HSL molecules from ''P. aeruginosa'', LasR binds to the '''[http://parts.igem.org/Part:BBa_J64010 LasI] promoter''' that controls the expression of the TEV protease, which we made available as [http://parts.igem.org/Part:BBa_K1319004 K1319004]. When the fusion protein is cleaved by the TEV protease, REACh will be separated from GFP. The latter will then be able to absorb and emit light as usual. |
{{Team:Aachen/Figure|align=center|Aachen_14-10-08_REACh_approach_with_BioBricks_iNB.png|title= Composition of our biosensor|subtitle=For our biosensor, we use a mix of already available and self-constructed BioBricks.|width=900px}} | {{Team:Aachen/Figure|align=center|Aachen_14-10-08_REACh_approach_with_BioBricks_iNB.png|title= Composition of our biosensor|subtitle=For our biosensor, we use a mix of already available and self-constructed BioBricks.|width=900px}} | ||
| - | The resulting fusion proteins | + | The resulting fusion proteins can be expressed in [http://parts.igem.org/Part:BBa_K1319013 K1319013] (GFP fused with REACh1) and [http://parts.igem.org/Part:BBa_K1319014 K1319014] (GFP fused with REACh2). The linker between the proteins containing a TEV protease cleavage site is labelled as [http://parts.igem.org/Part:BBa_K1319016 K1319016]. |
| + | |||
| + | Since we did not have enough time to build the complete system we tested our FRET reporter with an IPTG inducible TEV protease ([http://parts.igem.org/Part:BBa_K1319008 K1319008]). In this way we were able to establish a proof of concept of our reporter system including proper expression of the TEV protease as well as functionality of our GFP-REACh construct. | ||
| + | |||
| + | |||
| + | {{Team:Aachen/Figure|align=center|Aachen 14-10-17 IPTG REACh iFG.png|title= Composition of our biosensor for IPTG|subtitle=To test the funtionality of our sensor concept, we expressed the TEV protease with a T7 promoter.|width=900px}} | ||
| + | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 145: | Line 167: | ||
<span class="anchor" id="tevprotease"></span> | <span class="anchor" id="tevprotease"></span> | ||
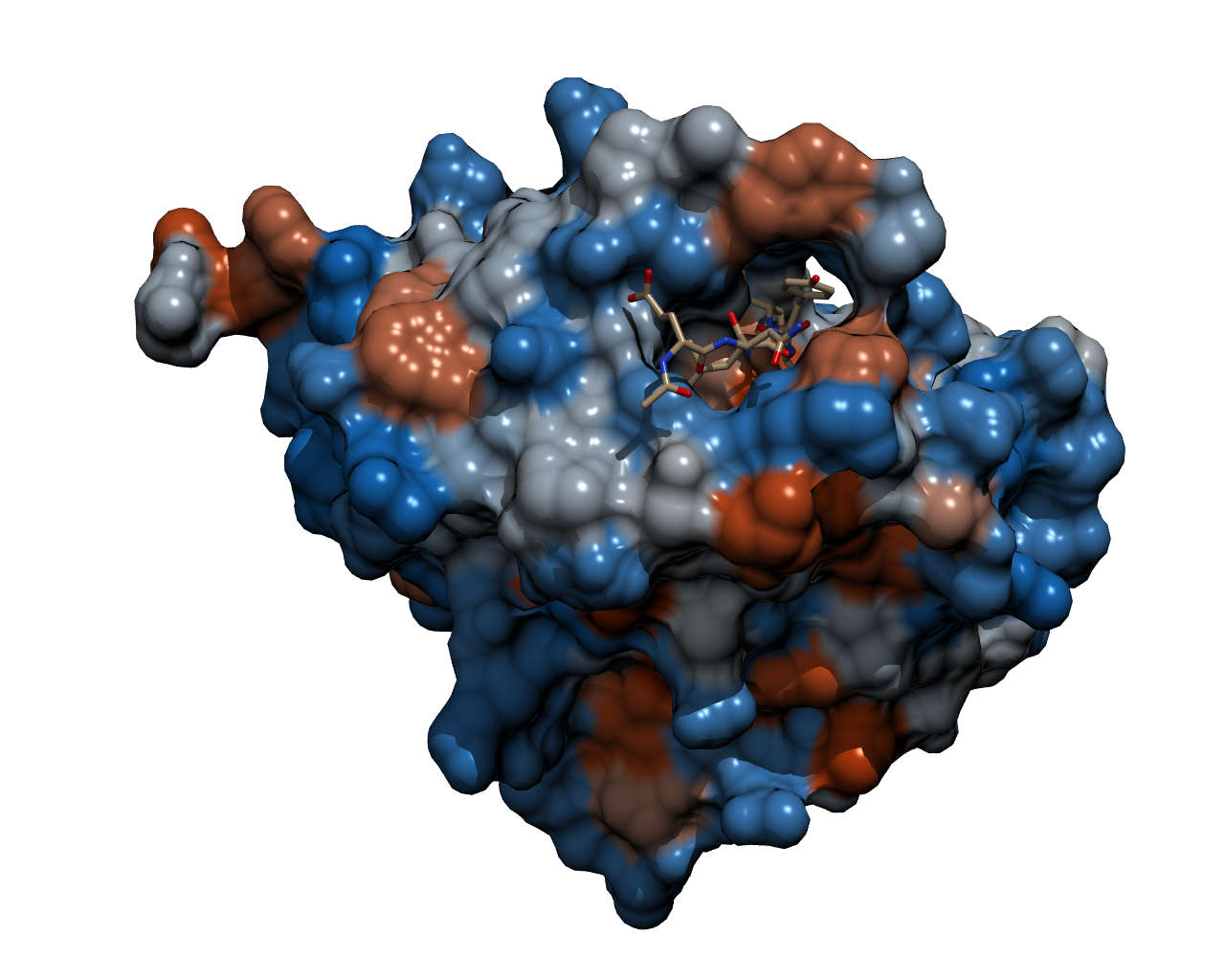
| - | To | + | To cleave the GFP-REACh fusion protein, we chose '''Tobacco Etch Virus (TEV) protease''', a highly sequence-specific cysteine protease, that is frequently used for the controlled cleavage of fusion proteins ''in vitro'' and ''in vivo''. The native protease also contains an internal self-cleavage site. This site is slowly cleaved to inactivate the enzyme. The physiological reason for the self-cleavage is unknown, however, undesired for our use. Therefore, our team uses a variant of the native TEV protease containing the mutation S219V which results in an alteration of the cleavage site so that self-inactivation is diminished. |
| - | {{Team:Aachen/Figure|Aachen_TEV_Protease_Model.png|title=TEV protease with a bound peptide|subtitle=This picture shows the TEV protease with a peptide chain bound in the binding pocket ready to be cleaved. The bound peptide chain | + | {{Team:Aachen/Figure|Aachen_TEV_Protease_Model.png|title=TEV protease with a bound peptide|subtitle=This picture shows the TEV protease with a peptide chain bound in the binding pocket ready to be cleaved. The recognition site of the bound peptide chain is located inside the binding pocket of the TEV protease. It was rendered with POV-Ray.|width=800px}} |
| + | |||
| + | Though quite popular in molecular biology, the TEV protease is not avaiable as a BioBrick yet. Hence, the team Aachen introduces the protease with anti-self cleavage mutation S219V and codon optimized for ''E. coli'' [http://parts.igem.org/Part:BBa_K1319004 '''to the Parts Registry this year.'''] | ||
| - | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| Line 155: | Line 178: | ||
[[File:Aachen_14-10-15_Medal_Cellocks_iNB.png|right|150px]] | [[File:Aachen_14-10-15_Medal_Cellocks_iNB.png|right|150px]] | ||
| - | = Achievements = | + | == Achievements == |
<span class="anchor" id="reachachievements"></span> | <span class="anchor" id="reachachievements"></span> | ||
| - | ==Characterization of GFP-REACh1 and GFP- | + | ===Characterization of GFP-REACh1 and GFP-REACh2 fusion proteins in combination with an IPTG-inducible TEV Protease=== |
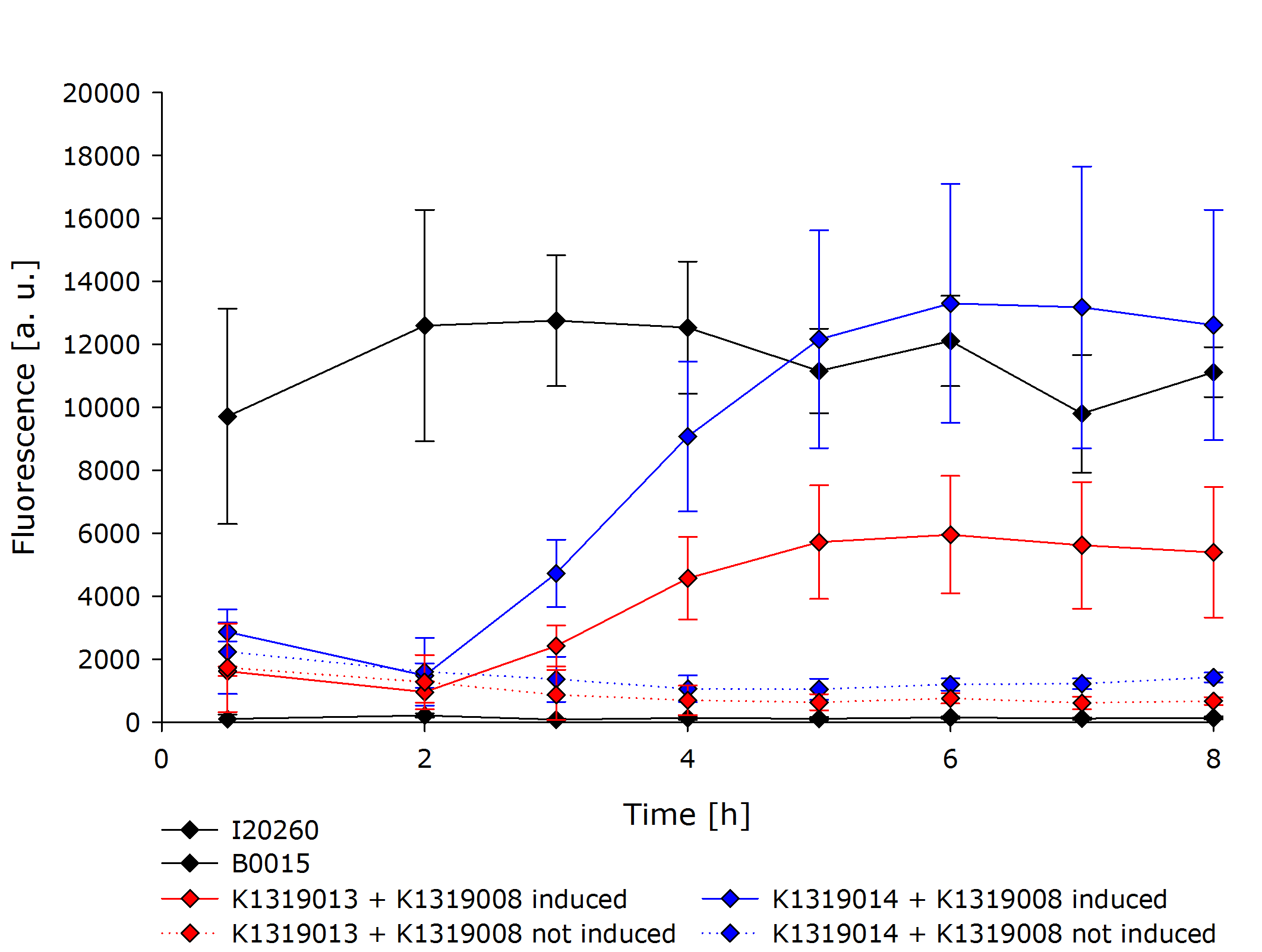
| - | The characterization of the TEV protease and the | + | The characterization of the TEV protease and the REACh1 and REACh2 dark quenchers was performed by introducing both simultaneously into ''E. coli'' Bl21 (DE3). The resulting double plasmid cells therefore contained [http://parts.igem.org/Part:BBa_K1319013 K1319013] (GFP-REACh1 fusion protein) or [http://parts.igem.org/Part:BBa_K1319014 K1319014] (GFP-REACh2 fusion protein) and [http://parts.igem.org/Part:BBa_K1319008 K1319008] (IPTG-inducible TEV protease). K1319013 and K1319014 were located on a pSB3K3 plasmid backbone and K1319008 on a pSB1C3 backbone, two standard iGEM plasmids with different oris allowing simultaneous use in one cell. |
| - | + | [http://parts.igem.org/Part:BBa_I20260 I20260] was used as a positive control because I20260 contains the same promoter ([http://parts.igem.org/Part:BBa_J23101 J23101]), the same RBS ([http://parts.igem.org/Part:BBa_B0032 B0032]) and the same version of GFP ([http://parts.igem.org/Part:BBa_E0040 E0040]) and is located on the same plasmid backbone pSB3K3. Therefore, it is expected that when all fusion proteins are successfully cut by the TEV protease, the fluorescence level of the double plasmid constructs reaches the same level as the positive control of I20260. As a negative control [http://parts.igem.org/Part:BBa_B0015 B0015] was used, a coding sequence of a terminator which should not show any sign of fluorescence. | |
| - | To | + | To characterize our REACh1/2 constructs in combination with the TEV protease a growth experiment was conducted. Both of the double plasmid constructs, a constitutive expression of GFP (I20260) as positive control and B0015 as negative control were compared. For each expression IPTG-induced and non-induced cultures were grown in parallel. All measurements were done in a biological triplicate. |
| - | + | To better evaluate the fluorescence, the observed Optical desity (OD) was taken into account in order to achieve a fluorescence measurement independent of the amount of cells present. This way, the measurement represents the amount of fluorescence per cell only. | |
| - | + | {{Team:Aachen/Figure|Aachen 16-10-14 Graph2iFG.PNG|title=Comparison of K1319013 + K1319008, K1319014 + K1319008, I20260 (positive control) and B0015 (negative control)|subtitle=Both double plasmid construct exhibit a clear fluorescence signal when induced.|width=700px}} | |
| - | + | The negative control B0015 did not exhibit any significant fluorescence. The positive control I20260 showed a steady level of fluorescence as expected due to the constitutive expression of GFP. As expected, the production is also independent of addition of IPTG therefore the triplicates have been merged together. | |
| - | + | Both double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 did not exhibit a strong fluorescence before induction with IPTG. In the non-induced state, the fluorescence stays low and does not increase over time. It is significantly weaker than the fluorescence reached by the induced constructs or the positive control but was higher than the negative control. The higher base level of fluorescence in the not induced constructs is due to the imperfect quenching and the leakiness of the IPTG inducible promoter. | |
| - | + | The induced double plasmid constructs exhibited a fast rise in fluorescence after induction. '''The signal strenght increased ~ 10-fold over the non-induced constructs.''' K1319014 + K1319008 reached the the same level of fluorescence as I20260, indicating a complete cleavage of the fusion proteins by the TEV protease. K1319013 + K1319008 did not reach a level of fluorescence as high as K1319013 + K1319008, however, the nearly 10-fold increase in fluorescence after induction is a clear indicator for the TEV protease cutting the fusion protein K1319013. The weaker fluorescence signal is probably due to a lower expression level of K1319013 in the cells compared to the expression level of K1319014. | |
| - | The | + | The fluorescence results of K1319014 + K1319008 was used to fit our [https://2014.igem.org/Team:Aachen/Project/Model model] to experimental data. |
| - | == | + | ====Summary==== |
| - | + | The double plasmid systems of K1319013 + K1319008 and K1319014 + K1319008 demonstrate the quenching ability of the REACh1 and REACh2 proteins as well as the funcionality of the TEV protease. | |
| + | The increase in fluorescence after induction with IPTG is based on a functional expression of the TEV protease which proceeds to cut the linker of the fusion protein already produced destroying the FRET system between GFP and its quencher and resulting in a strong fluorescence signal. Combined, this characterization is a '''validation of the functionality of the REACh1 protein ([http://parts.igem.org/Part:BBa_K1319001 K1319001]), the REACh2 protein ([http://parts.igem.org/Part:BBa_K1319002 K1319002]) and the TEV protease ([http://parts.igem.org/Part:BBa_K1319004 K1319004])'''. | ||
| - | + | ===Determining the Quenching ability of REACh1 and REACh2=== | |
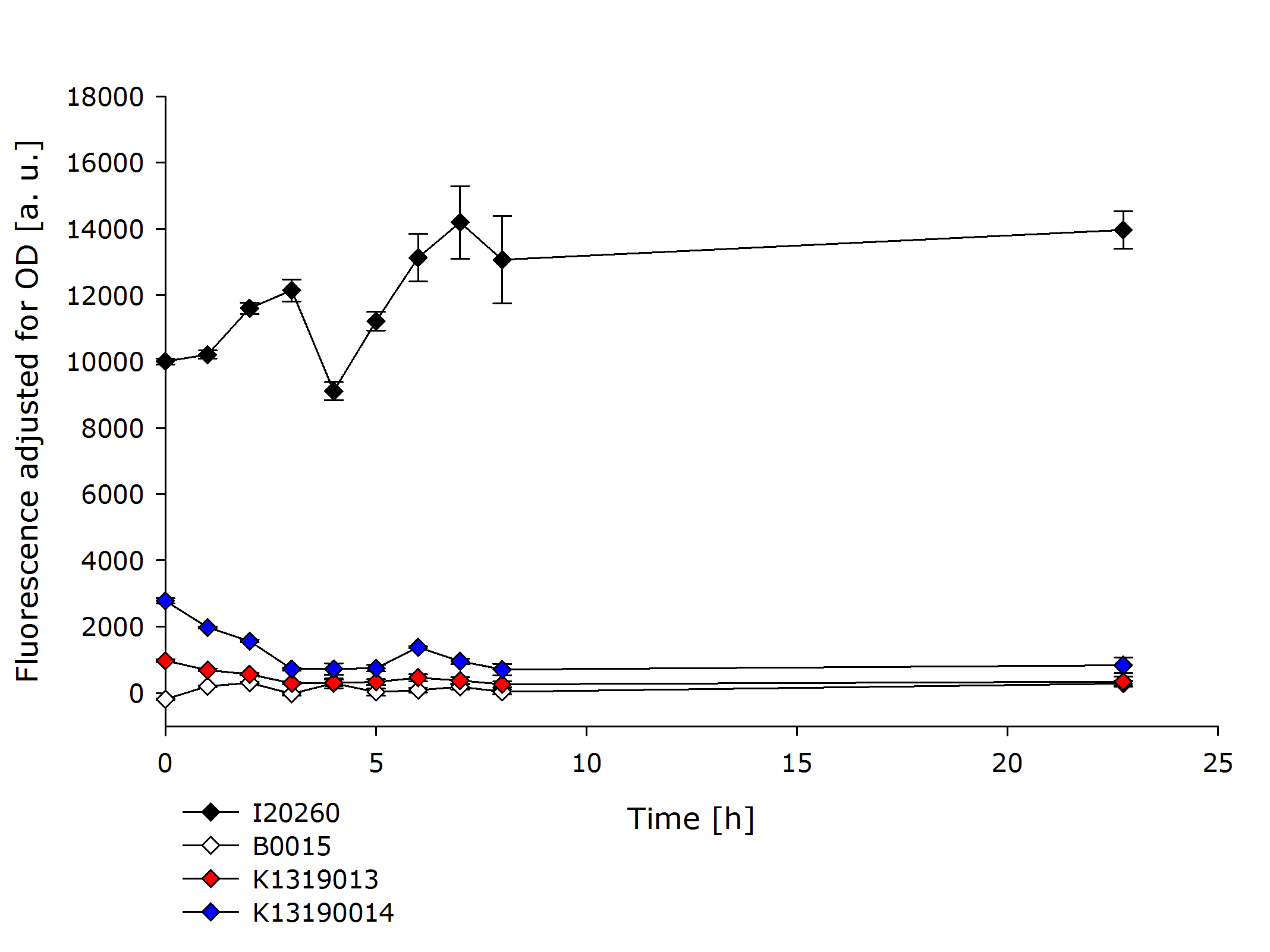
| - | + | In order to further evaluate the quenching ability of the REACh1 and REACh2 constructs in the fusion proteins produced by K1319013 and K1319014, they were expressed alone without an IPTG inducible TEV protease. This eliminated the effect of a potential leakiness of the non induced promoter to reliably assess the quenching ability of the REACh1 and REACh2 proteins. | |
| + | {{Team:Aachen/Figure|Aachen_K1319001_and_K1319002.PNG|title=Comparison of K1319013 and K1319014 with I20260 and B0015|subtitle=K1319013 and K1319014 show a severely reduced fluorescence compared to the positive control I20260.|width=700px}} | ||
| - | + | In the previous experiment it was established that the fusion proteins K1319013 and K1319014 are expressed funtionally. K1319014 reached the same level of fluorescence as the positive control after being cut by the TEV protease. Therefore the reduced fluorescence in this experiment is completely attributable to the quenching of REACh1. The quenching reduces the fluorescence of GFP by a factor ~ 25 which means a '''quenching efficiency of ~ 96%!'''. | |
| - | + | K139013 was not able to reach the same fluorescence level as the positive control in the previous experiment. Therefore a worse rate of functional expression of K1319013 compared to K1319014 is assumed. incorporating this, the difference in fluorescence between induced and not induced is still a factor of ~ 30 resulting in a '''quenching efficiency of ~ 97%'''. | |
| - | + | ====Summary ==== | |
| - | + | The fusion proteins of GFP combined with REACh1 and REACh2 are not only fully functional but exhibit a great quenching efficiency of ~ 97% for REACh1 and ~ 96% for REACh2.Even though REACh1s quenching ability seems to be slighty superior, the expression level of the K1319014 fusion protein including REACh2 is nearly twice as high as the fusion protein K1319013 containing REACh1 and therefore shows a stronger fluorescence. Ganesan et al. (2006) reported a reduction on emission of GFP of 82% for REACh1 and 98% for REACh2 but with a different Linker between the proteins and on a different vector backbone. | |
| - | ''''' | + | ===Comparing fluorescence kinetics of the GFP-REACh fusion proteins with a Standard lacI-inducible GFP Expression=== |
| + | |||
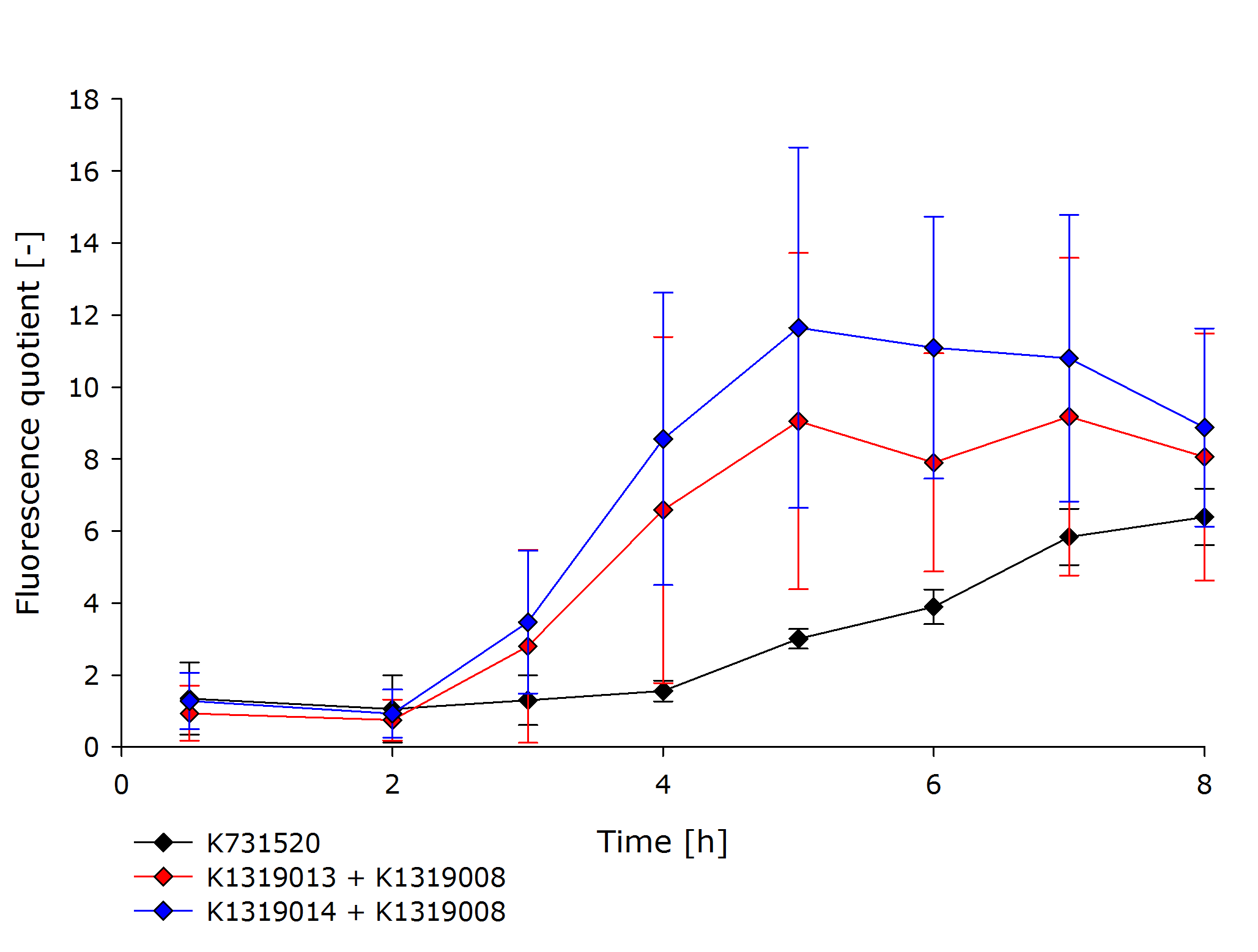
| + | To assess the kinetics of the fusion proteins K1319013 (GFP-REACh1) and K1319014 (GFP-REACh2), the double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 were compared to a standard expression of GFP under the control of a lacI promoter in [http://parts.igem.org/Part:BBa_K731520 K731520], a BioBrick made by the iGEM Team TRENTO in 2012. This tested the hypothesis of achieving a faster fluorescence response with the GFP-REACh fusion proteins compared to a standard expression. | ||
| + | |||
| + | K731520 and the double plasmid constructs K1319013 + K1319008 and K1319014 + K1319008 were cultivated in ''E. coli'' BL21(DE3), and fluorescence and OD was measured.The fluorescence was adjusted for the OD to show a relative fluorescence on a per cell basis. The difference between the induced and non-induced state, the fluorescence quotient, serves as a better indicator for a system used as a sensor because the difference between an ''on'' and ''off'' state is more important for a clear and unmistakable signal than the overall fluorescence. Hence, the OD-adjusted fluorescence quotient for both double plasmid constructs and K731520 was obtained and plotted in the following graph. | ||
| + | |||
| + | {{Team:Aachen/Figure|Aachen_16-10-14_GraphQuotient_iFG.PNG|Comparison of K1319013 + K1319008, K1319014 + K1319008 and K731520|subtitle=Fluorescence was normalized to the cell growth. The fluorescence of induced cells was additionally divided by the fluorescence of non-induced cells to obtain the fluorescence quotient.|width=700px}} | ||
| + | |||
| + | The graph clearly shows the faster response of the cut GFP-REACh fusion protein compared to a standard GFP expression. Both fluorescence signals of the double plasmid constructs achieve a higher difference in fluorescence signal between induced and non-induced state as well as at a faster rate. This proves the hypothesis made earlier about the kinetics of the GFP-REACh fusion protein combined with the TEV protease. | ||
| + | |||
| + | ====Summary==== | ||
| + | |||
| + | The kinetics of the fusion protein combined with the TEV protease exhibits the exact characteristics as predicted. The response is clearly faster than normal expression by accumulating a reservoir of fusion proteins which are not fluorescing due to the dark quencher attached to them. This reservoir is then activated by the induction of the TEV protease expression. Production of the protease results in the cleavage of the fusion protein, releasing GFP from the dark quencher and disturbing the interaction between the FRET pair. This results in the observed faster fluorescence reaction due to the amplificating effect of the TEV protease in which every one TEV protease can account for many fluorescence proteins being activated. | ||
| + | |||
| + | ===Characterizing the GFP-REACh Constructs in Sensor Chips=== | ||
| + | To further characterize the REACh construct, they were introduced into the sensor cells which were then induced with 2 µL IPTG with a concentration of 100 mM. Subsequently, we took fluorescence measurement read-outs (GFP, excitation 496 ± 9 nm, emission 516 ± 9 nm) roughly every 10 min in the plate reader. The results were plotted in the heatmap shown here. | ||
| + | <center> | ||
| + | <div class="figure" style="float:{{{align|center}}}; margin: 0px 10px 10px 40px; border:{{{border|0px solid #aaa}}};width:{{{width|500px}}};padding:10px 10px 0px 0px;"> | ||
| + | {| | ||
| + | |<html> <img src="https://static.igem.org/mediawiki/2014/6/6d/Aachen_K1319014%2B8_and_K1319013%2B8_pixelig_minus_bg.gif" width="500px"></html> | ||
| + | |- | ||
| + | |'''{{{title|K1319014 + K1319008 and K1319013 + K1319008 non-induced (top) and induced (bottom) in sensor cells }}}'''<br />{{{subtitle|The induced double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 exhibit a clear fluorescence response in our sensor cells which in response to induction with 2 µl IPTG. }}} | ||
| + | |} | ||
| + | </div> | ||
| + | </center> | ||
| + | |||
| + | The heatmap shows an increase of fluorescence from blue (no fluorescence) to red (high fluorescence). It is clearly visible that the induced chips are exhibiting a significantly higher fluorescence than the non-induced chips. This shows that the constructs also work as intended in the sensor chips: The TEV protease cuts the linker so that the fusion protein is separated into GFP and a dark quencher, disabling the quenching. GFP has a clear fluorescence emission after the fusion protein has been successfully cut into two pieces by the TEV protease. | ||
| + | |||
| + | ===Comparing the kinetic of the double plasmid systems K1319013 + K1319008 and K1319014 + K1319008 with standard GFP expression=== | ||
| + | |||
| + | <center> | ||
| + | <div class="figure" style="float:{{{align|center}}}; margin: 0px 10px 10px 40px; border:{{{border|0px solid #aaa}}};width:{{{width|480px}}};padding:10px 10px 0px 0px;"> | ||
| + | {| | ||
| + | |<html> <img src="https://static.igem.org/mediawiki/2014/1/1a/Aachen_K13%2B8%2CK14%2B8%2CK731_slower_reduced.gif" width="480px"></html> | ||
| + | |- | ||
| + | |'''{{{title|K1319013 + K1319008, K1319014 + K1319008 and K731520 in an non-induced (top) and induced (bottom) chip}}}'''<br />{{{subtitle|Comparing the factor of fluorescence increase of induced (bottom) and not induced (top) sensor chips to the first frame of the measurement. The tested constructs are K1319013 + K1319008, K1319014 + K1319008 and K731520. After five hours, the fluorescence in our dual-plasmid REACh construct contaning K1319014 had increased by a factor of about 8, while K731520 fluorescence increased by only a factor of 2-3.}}} | ||
| + | |} | ||
| + | </div> | ||
| + | </center> | ||
| + | |||
| + | The analysis of the different [https://2014.igem.org/Team:Aachen/Project/2D_Biosensor sensor chips] with the three different construct K1319013 + K1319008, K1319014 + K1319008 and K731520 demonstrates the same fluorescence response kinetic as in the shake flask experiments. The double plasmid systems exhibits a faster and stronger fluorescence response compared to a standard GFP expression in K731520. The build up pool of fusion proteins allows for a faster, stronger fluorescence response when the induced TEV protease cleaves the fusion proteins and releases GFP from its dark quencher. | ||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
| - | + | [[File:Aachen_14-10-16_Outlook_Cellocks_iNB.png|right|150px]] | |
| - | + | == Outlook == | |
| + | <span class="anchor" id="reachoutlook"></span> | ||
| - | + | The system of the GFP-REACh fusion proteins with an inducible TEV protease has been established and shows the desired results of being faster than standard expression. The next step will be to devise an expression of the TEV protease inducible by HSL instead of IPTG and then to incorporate both the HSL inducible TEV protease and the fusion protein into one plasmid backbone. This would also allow us to choose a high copy plasmid for both inserts, instead of a high copy plasmid for the TEV protease and a low to mid copy plasmid for the fusion protein which should yield an overall higher fluorescence readout. | |
| + | |||
| + | Subsequently the combined construct will be characterizes the same way as the double plasmid system and other options regarding different fluorescence proteins with different quenchers will be considered to be able to have multiple fluorescence responses readable at the same time while still being faster than normal expression. | ||
| + | |||
| + | Also finding and testing different promoters which can be used to express the TEV protease is planned to be able to detect not only ''Pseudomonas aeruginosa'' but also other pathogens. Also the technology can be expanded to other molecules enabling other research areas to profit from the faster fluorescence response. | ||
| - | |||
{{Team:Aachen/BlockSeparator}} | {{Team:Aachen/BlockSeparator}} | ||
= References = | = References = | ||
| + | * Sundar Ganesan, Simon M. Ameer-Beg, Tony T. C. Ng, Borivoj Vojnovic and Fred S. Wouters. "A dark yellow fluorescent protein (YFP)-based Resonance Energy-Accepting Chromoprotein (REACh) for Förster resonance energy transfer with GFP" Proceedings of the National Academy of Science of the United States of America | March 14,2006 | vol. 103 | no. 11 | 4089 - 4094 | ||
| + | |||
* Broussard, Joshua A, Benjamin Rappaz, Donna J Webb, and Claire M Brown. "Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt." Nature protocols 8.2 (2013): 265-281. doi:10.1038/nprot.2012.147. | * Broussard, Joshua A, Benjamin Rappaz, Donna J Webb, and Claire M Brown. "Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt." Nature protocols 8.2 (2013): 265-281. doi:10.1038/nprot.2012.147. | ||
| - | * | + | * Broussard, J. A., Rappaz, B., Webb, D. J., & Brown, C. M. (2013). Fluorescence resonance energy transfer microscopy as demonstrated by measuring the activation of the serine/threonine kinase Akt. Nature protocols, 8(2), 265-281. doi: 10.1073/pnas.0509922103 |
'''SWISS-MODEL''' | '''SWISS-MODEL''' | ||
| - | * | + | * Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2005). The SWISS-MODEL Workspace: A Web-based Environment For Protein Structure Homology Modelling. Bioinformatics, 22(2), 195-201. |
| - | + | ||
| - | * | + | * Biasini, M., Bienert, S., Schwede, T., Waterhouse, A., Arnold, K., Studer, G., et al. (2014). Nucleic Acids Research. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. doi: 10.1093/nar/gku340. |
| - | * Guex, N., Peitsch, M.C., Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 30(S1), S162-S173. | + | |
| + | * Guex, N., Peitsch, M. C., & Schwede, T. (2009). Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis, 30(S1), S162-S173. | ||
| + | |||
| + | * Kiefer, F., Arnold, K., Kunzli, M., Bordoli, L., & Schwede, T. (2009). The SWISS-MODEL Repository and associated resources. Nucleic Acids Research, 37(Database), D387-D392. | ||
'''UCSF Chimera''' | '''UCSF Chimera''' | ||
| - | * Pettersen | + | * Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera?A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-1612. PubMed PMID: 15264254. |
'''POV-Ray''' | '''POV-Ray''' | ||
* Persistence of Vision Pty. Ltd. (2004) Persistence of Vision Raytracer (Version 3.7) [Computer software]. Retrieved from http://www.povray.org/download/ | * Persistence of Vision Pty. Ltd. (2004) Persistence of Vision Raytracer (Version 3.7) [Computer software]. Retrieved from http://www.povray.org/download/ | ||
| + | |||
{{Team:Aachen/Footer}} | {{Team:Aachen/Footer}} | ||
Latest revision as of 03:47, 18 October 2014
|
|
|
|
|
|
|
|
 "
"