|
Achievements
Medium
Prior to using our own device for detection of fluorescence emitted by the sensor chips we used equipment readily available in the lab. A Molecular Imager® Gel DocTM XR+ from BIO-RAD was available which used UV and white light illuminators. Only two different filters were availble for the excitation ligth wavelength, which resulted in very limitted possibilities for excitation of fluorescent molecules. For example, it was possible to detect the expression of iLOV in our sensor chips, but in contrast detection of GFP was not possible. Thus the Gel DocTM XR+ was not ideal for our project.
(iLOV_GFP_HM_1,5h.png)
Concerning the medium used for our sensor chips, LB medium showed a high background fluorescence when exposed to UV light. Surprinsingly the background fluorescence resulting from the LB medium was to high to detect a signal emitted by our sensor cells, thus we used minimal media (NA, M9, Hartman) in order to minimize background fluorescence. The appliction of minimal media was sufficient to minimize the background fluorescence, but this approach came with the drawback of minimal to zero growth of our sensor cells.
(Chip_medium_geldoc.png)
Further experiments were conducted concerining long-time storage of the sensor chips. Storage at -20 °C resulted in the loss of our sensor cells. Adding 5-10% glycerol ensured survival of the sensor cells, but resulted in an expression stop of fluorescence proteins. Thus the idea of long time storage of the sensor chips hab to be rejected. However, it was possible to store readily usable sensor chips for 2 days at 4 °C when using LB medium and storage for 5 days was possible, if TB medium was used.
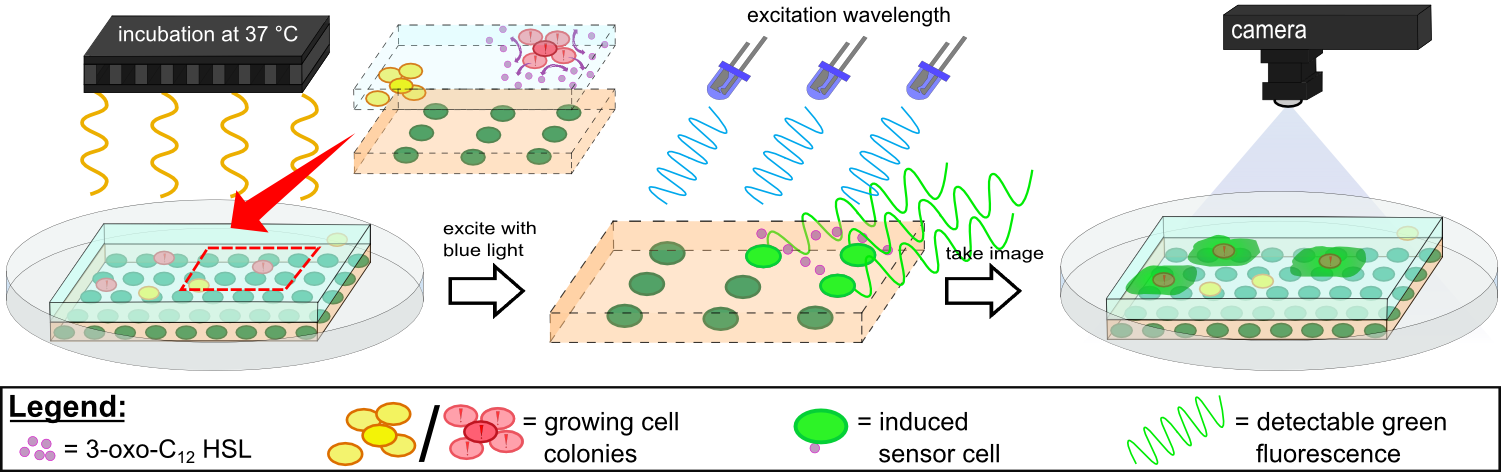
In our own device WatsOn optimized wavelengths of 450 nm and 480 nm were used for excitation of iLOV and GFP, respectively. When exposed to both excitation wavelengths the LB medium showed minimal background fluorescence and no difficulties were observed in cultivation of our sensor cells. Further reduction of background fluorescence compared to LB medium was observed when using TB medium for sensor chip manufacturing in combination with fluorescence evaluation using WatsOn.
(5Tage_K131026_neb_tb_1,5h)
Agar concentration
For sensor chip manufacturing an optimal agarose concentration of 1.5% was found. When agarose concentrations below 1.5% (w/v) were used the sensor chips were easily damaged and were not transportable. Agar concentrations above 1.5% (w/v) had to be avoided, because the agarose started to solidify before it could be poured into the chip mold.
Agarose was chosen above agar, because of more evely linkage and thus more homogenous chips. In addition agarose reduced diffusion of inducer molecules through the chip. Reduced diffusion was desired in order to achieve distinct fluorescent spots on the sensor chips.
Chip form
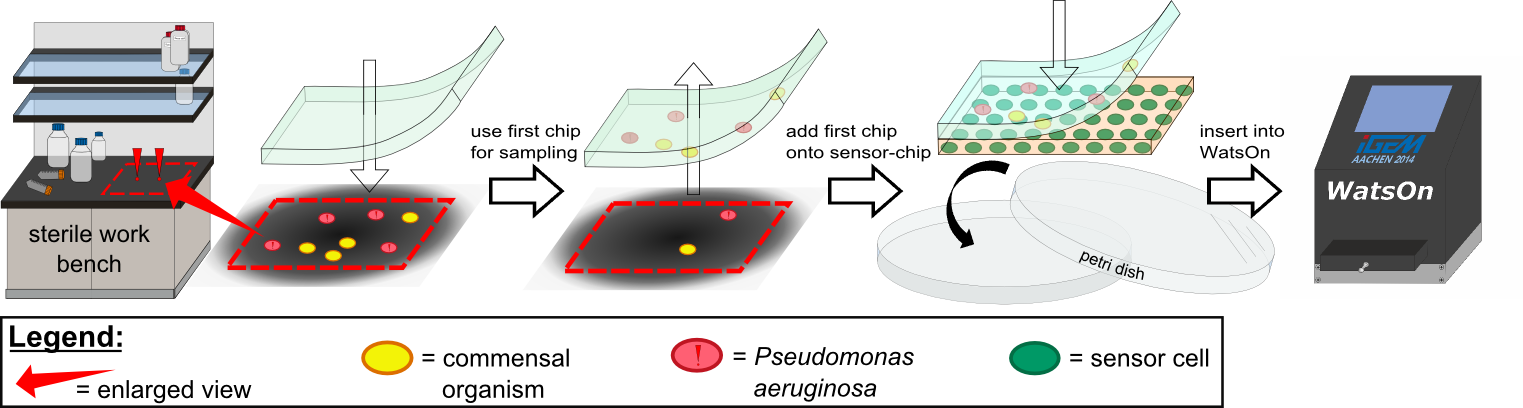
Various approaches were tried for production of sensor chips with reproducable quality. The first approach was to cast every sensor chip indicidually. In order to archive a plain chip surface, which was required for high quality images, we tried to cast the sensor chips between two microscope slides. This approach had to be rejected, because the agar was to liquid. Second, we produced a closed mold in which the liquid agar was injected using a pipette, but we encountered a high frequency of bubbles in the chips when using this approach.
(2_chipform.jpg)
In the end we use a self made open form. Here you produce 9 chips at the same time. You just have to cut them out (anhand) the lasered lines. This is a fast, easy and efficient way to produce chips. And because of the surface tension the chips are plane.
(final_chipform.jpg)
Induction
For the induction of the used constructs we use IPTG or 3-oxo-C12 HSL.
The sensor cells with K1319042 in BL21 can detect a IPTG concentration of 1 mM (0,2 µl).
(2µl_IPTG_1mM_K1319042_1h.png)
The sensor with the REACh constructs in BL21 can detect a IPTG concentration of ???
(Plate reader???)
The sensor cells with K131026 in BL21 can detect an HSL concentration of 500 µg/ml (0,2 µl).
It also can detect Pseudomonas aeruginosa.
(Zeitaufnahmen bearbeitet von Arne)
Testing our sensor Chips with a Platereader
Detecting the 3-oxo-C12 HSL with K131026 in our sensor chip with WatsOn.
Detecting Pseudomonas aeruginosa with K131026 in our sensor chip with WatsOn.
|
 "
"