Team:LMU-Munich/Team/Collaborations
From 2014.igem.org
(→The cloning:) |
(→Flow through cytometry analysis:) |
||
| Line 23: | Line 23: | ||
=== Flow through cytometry analysis: === | === Flow through cytometry analysis: === | ||
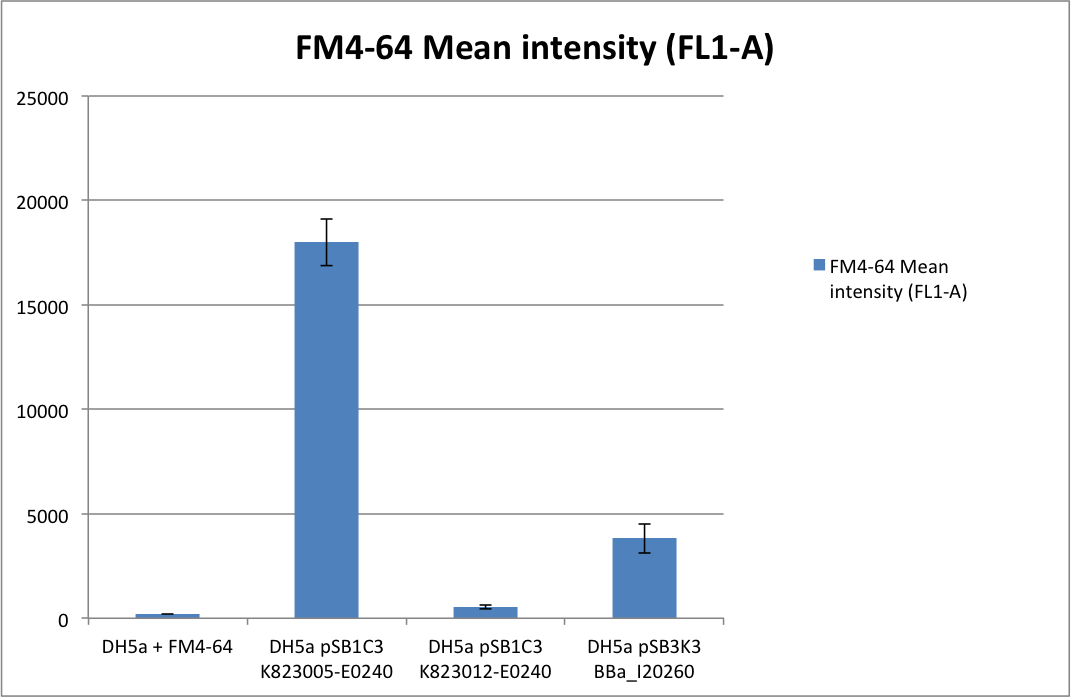
For one measurement 50 µl of an overnight culture was first incubated together with 1 µl FM4-64 at 37°C to later on identify life/dead cells. After that 1 µl of each sample was diluted in 200 µl phosphate buffered saline solution and then evaluated by flow through cytometry. In order to generate meaningful results the described measurement was perfomed three times with technical replicas. For complete details please check our Interlab study submission: [[Media:LMU14_Interlab_3_091014.pdf | LMU-Munich Interlab form]]<br> | For one measurement 50 µl of an overnight culture was first incubated together with 1 µl FM4-64 at 37°C to later on identify life/dead cells. After that 1 µl of each sample was diluted in 200 µl phosphate buffered saline solution and then evaluated by flow through cytometry. In order to generate meaningful results the described measurement was perfomed three times with technical replicas. For complete details please check our Interlab study submission: [[Media:LMU14_Interlab_3_091014.pdf | LMU-Munich Interlab form]]<br> | ||
| - | + | ||
=== Results: === | === Results: === | ||
<br> | <br> | ||
Revision as of 18:36, 11 October 2014
Collaborations
Collaborations and the exchange of knowledge were important aspects during our iGEM participation. Therefore we had a intensive collaboration with the iGEM team Groningen (Netherlands) and laid a high priority on the exchange with the Rathenau Institute also located in the Netherlands. In addition we also took part in the Interlab Study of this year´s iGEM competition.
Interlab Study
Background:
The aim of the Interlab study was to evaluate three devices consisting of different Anderson promoters fused to GFP in two different backbones: pBS1C3 and pBS3K3. The devices were distributed by iGEM and the flourescence evaluation method was free of choice. For more details check: Interlab Study@iGEM
The devices:
- Device one: BBa_I20260 consisting of J23101-B0032-E0040-B0015 in the backbone pBS3K3
- Status: ready for evaluation
- Device two: BBa_J23101 + BBa_E0240 in the backbones pBS1C3
- Status: cloning before evaluation
- Device three: BBa_J23115 + BBa_E0240 in the backbones pBS1C3
- Status: cloning before evaluation
The cloning:
Devices two and three were built from their individual parts. By digesting the promotors (BBa_J23101and BBa_J23115) with SpeI-HF and PstI-HF restriction enzymes the backbone was cut open. The XbaI and PstI-HF digested GFP (BBa_E0240) was then ligated behind the promotors. After that both devices were transformed into DH5α, plated out on chloramphenicol [35mg/µl] LB-agar plates and tested via colony-PCR to identify positive clones. For clarification PCR-positive clones were sequenced: BBa_J23101 + BBa_E0240.txt BBa_J23115 + BBa_E0240.txt
Flow through cytometry analysis:
For one measurement 50 µl of an overnight culture was first incubated together with 1 µl FM4-64 at 37°C to later on identify life/dead cells. After that 1 µl of each sample was diluted in 200 µl phosphate buffered saline solution and then evaluated by flow through cytometry. In order to generate meaningful results the described measurement was perfomed three times with technical replicas. For complete details please check our Interlab study submission: LMU-Munich Interlab form
Results:
Discussion:
Collaboration with Team Virginia
Collaboration with Team Groningen

Hi there!
Welcome to our Wiki! I'm BaKillus, the pathogen-hunting microbe, and I'll guide you on this tour through our project. If you want to learn more about a specific step, you can simply close the tour and come back to it anytime you like. So let's start!

What's the problem?
First of all, what am I doing here? The problem is, pathogenic bacteria all around the world are becoming more and more resistant against antimicrobial drugs. One major reason for the trend is the inappropriate use of drugs. With my BaKillus super powers, I want to reduce this misuse and thus do my part to save global health.

Sensing of pathogens
To combat the pathogenic bacteria, I simply eavesdrop on their communication. Bacteria talk with each other via quorum sensing systems, which I use to detect them and trigger my responses.

Adhesion
The more specific and effective I can use my powers, the lower the danger is of provoking new resistance development. So I catch pathogens whenever I get hold of them and stick to them until my work is done.

Killing
Talking about my work - killing pathogens is finally what I am made for. In response to quorum sensing molecules of the pathogens, I export a range of antimicrobial substances leading to dissipation of biofilms and the killing of the targeted bacteria.

Suicide switch
When the job is done and all the bad guys are finished, you don't need a super hero anymore. So after fulfilling my work I say goodbye to the world by activating my suicide switch.

Application
Of course I'm not only a fictional hero, but a very real one. In two different prototypes, I could be used for diagnosis or treatment of pathogen-caused diseases. However, there is still a whole lot of regulational and economical questions that have to be answered before.

See you!
So now you know my short story - and it is time for me to return to my fight for a safer world. Feel free to take a closer look on my super powers, the process of my development or the plans for a medical application.
 "
"