Team:SUSTC-Shenzhen/Notebook/CRISPR/Follow-up-for-verification-of-plasmid-PX330-MCherry-GAL4
From 2014.igem.org
Notebook
Elements of the endeavor.
Contents |
Follow-up for verification of plasmid PX330-MCherry-GAL4 and PX330-MCherry and The results of ‘struggles for fill-in(the second time)
2014/8/12 Verification for plasmidsFollow-up for verification of plasmid PX330-MCherry-GAL4、PX330-MCherry (amplified on 2014.8.9) by restriction digestion
We decided to repeat the verification experiment for plasmid MCherry-GAL4-PX330、MCherry-PX330, since we had failed yesterday.
Methods
The same as yesterday’s verification experiment
Methods
Perform restriction digestion for plasmids MCherry-PX330(MCherry2-1、2-2、2-3, MCherry-GAL4-PX330(1-1、1-4)
Digestion system: [unit: ul]
| DNA | EcoRI | Age I | Buffer | ddH2O | Total volume | |
| MCherry2-1(188.5) | 0.5 | 1 | 1 | 1 | 6.5 | 10 |
| MCherry2-2(229.6) | 0.5 | 1 | 1 | 1 | 6.5 | 10 |
| MCherry2-3(196.2) | 0.5 | 1 | 1 | 1 | 6.5 | 10 |
| GAL4 1-1(239.0) | 0.5 | 1 | 1 | 1 | 6.5 | 10 |
| GAL4 1-4(201.1) | 0.5 | 1 | 1 | 1 | 6.5 | 10 |
Incubate at 37°C for 4 hours
Loading system: [unit: ul]
| DNA | Dye | Total volume | |
| MCherry(2-1、2-2、2-3)\GAL4(1-1、1-4) | 9.5 | 2.5 | 12 |
Running conditions: 120V, 45min
Results of electrophoresis:
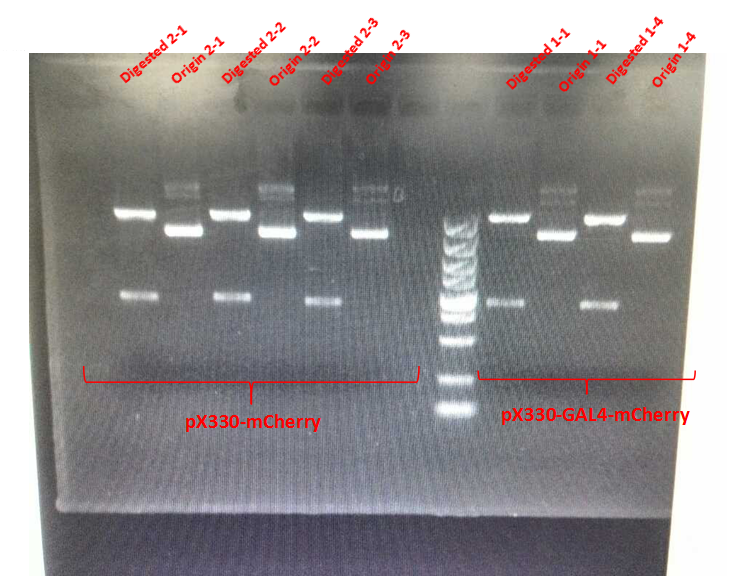
From the picture, we can see that, for those digested group, both of the two supposed bands (4200, 800bp) had appeared, which meant that the amplification for plasmids pX330-mCherry and pX330-GAL4-mCherry had succeeded.
The results of ‘struggles for fill-in(the second time)’
Results of plasmid Purification:
| Concentration(unit:ng/μL) | |
|---|---|
| G12-1 | 26.5 |
| G12-2 | 41.0 |
| G34-1 | 63.6 |
| G34-2 | 51.9 |
| G56-1 | 35.6 |
| polyA56-1 | 26.1 |
| polyA56-2 | 23.3 |
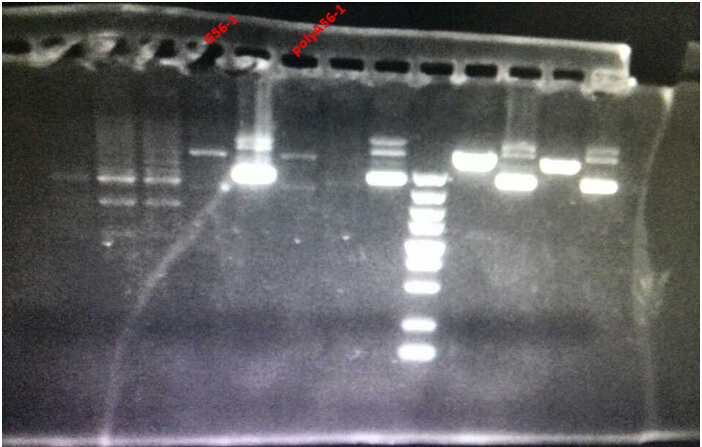
From the picture, we can see that, G56-1and polyA56-1 only suffered one cut during the digestion, which implied the slight possibility of the success for Fill-in ( EcoRI site has been eliminated by fill-in). Thus, we decided to perform further verifications for G56-1and polyA56-1.
 "
"