Team:Paris Saclay/Notebook/August/6
From 2014.igem.org
(→PCR Cleanup) |
m (→Wednesday 6th August) |
||
| (41 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Paris_Saclay/notebook_header}} | ||
=Wednesday 6th August= | =Wednesday 6th August= | ||
==Lab work== | ==Lab work== | ||
| - | === | + | ===The chassis coli Odor free=== |
| - | ====PCR | + | ====Final stock of MG1655==== |
| + | ''by Romain'' | ||
| + | |||
| + | 2 final stocks of the MG1655 made with 1ml of strain and 250µl glycerol 87%, in -80°C. | ||
| + | |||
| + | ===Salicylate Inducible Suppressing System=== | ||
| + | ====Plasmid DNA Purification==== | ||
| + | ''by Eugene and Fabio'' | ||
| + | * BBa_K1372000 Cl.1 | ||
| + | * BBa_K1372000 Cl.2 | ||
| + | From Liquid Culture made the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/5#C_-_Salicylate_Inducible_Suppressing_System 5th August] | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/Extraction_of_the_Genomic_DNA_from_Bacteria_by_using_NucleoSpin®_Tissue Protocol] | ||
| + | |||
| + | ====Digestion==== | ||
| + | ''by Eugene and Fabio'' | ||
| + | |||
| + | In order to place the '''BBa_B0015''' just after the '''BBa_K1372000''', we will start the [http://parts.igem.org/Help:Assembly/3A_Assembly Assembly procedure] again. | ||
| + | * BioBrick '''BBa_K1372000''' (Salicylate promoter + NahR + RNA suppressor) as '''Part A''' | ||
| + | * BioBrick '''BBa_B0015''' (Double terminator) as '''Part B''' | ||
| + | The enzymes used were: | ||
| + | |||
| + | * '''EcoRI''' and '''SpeI''' for '''BBa_K1372000''' | ||
| + | * '''EcoRI''' and '''XbaI''' for '''BBa_B0015''' | ||
| + | |||
| + | <font color="#F00">TODO: Illustration of the process</font> | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/BioBrick_Assembly#Digest_reaction BioBrick Assembly - Digest Reaction Protocol] | ||
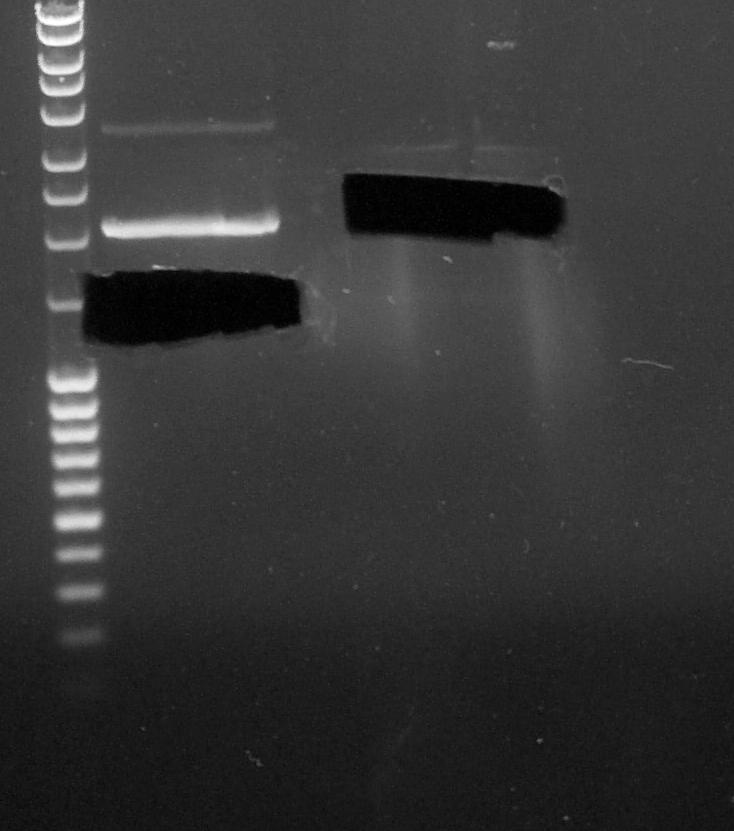
| + | ====Segregate Process by Electrophoresis==== | ||
| + | |||
| + | [[File:Paris_Saclay_060814_cut_gel.jpg|200px|left]] | ||
| + | |||
| + | ''by Eugene and Fabio'' | ||
| + | |||
| + | # BBa_B0015 digested by '''EcoRI''' and '''XbaI''' | ||
| + | # BBa_K1372000 digested by '''EcoRI''' and '''SpeI''' | ||
| + | |||
| + | |||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/BioBrick_Assembly#Segregate_process BioBrick Assembly - Segregate Process Protocol] | ||
| + | |||
| + | ===Lemon scent=== | ||
| + | ====PCR Clean-up==== | ||
''by Pierre & Sean'' | ''by Pierre & Sean'' | ||
| + | |||
| + | BioBricks used: '''BBa_K762100''' and '''BBa_K517003''' (PCR perfomed on the [https://2014.igem.org/Team:Paris_Saclay/Notebook/August/4 4th August]) | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/PCR_clean-up Protocol] | ||
| + | |||
| + | ====PCR digestion==== | ||
| + | ''by Pierre & Sean'' | ||
| + | |||
| + | |||
| + | '''Protocol''' | ||
| + | |||
| + | For each BioBrick ('''Bba_K517003''' and '''BBa_K762100''')mix the following into a microcentrifuge tube. | ||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | component | ||
| + | ! scope=col | volume | ||
| + | |- | ||
| + | |H<sub>2</sub>O | ||
| + | |5.5μl | ||
| + | |- | ||
| + | | PacI | ||
| + | | .5μl | ||
| + | |- | ||
| + | | Fast Digest buffer | ||
| + | | 4μl | ||
| + | |- | ||
| + | | Cleaned-up PCR content | ||
| + | | 30μl | ||
| + | |} | ||
| + | |||
| + | Then bain-marie at 37°C for one hour. | ||
| + | Finally conduct PCR cleanup for both BioBricks, with 15µl of NE buffer. | ||
| + | |||
| + | ''We acidentally used PvuI instead of PacI. - SC 6/8/14'' | ||
| + | |||
| + | ====PCR pCola plasmide (Geranyl Synthase)==== | ||
| + | ''by melanie'' | ||
| + | |||
| + | amplification of geranyl Synthase | ||
| + | |||
| + | [https://2014.igem.org/Team:Paris_Saclay/Protocols/PCR_for_the_genomic_DNA_of_bacteria protocol] | ||
| + | |||
| + | PCR protocol: | ||
| + | 98° --> 2min | ||
| + | |||
| + | 5 PCR cycle: | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | time | ||
| + | ! scope=col | 10sec | ||
| + | ! scope=col | 20sec | ||
| + | ! scope=col | 45sec | ||
| + | |- | ||
| + | !temperature | ||
| + | |98° | ||
| + | |(depending on the tube - see the gradient) | ||
| + | |72° | ||
| + | |} | ||
| + | |||
| + | 25 PCR cycle | ||
| + | |||
| + | {| class="wikitable centre" width="50%" | ||
| + | |+ | ||
| + | |- | ||
| + | ! scope=col | time | ||
| + | ! scope=col | 10sec | ||
| + | ! scope=col | 20sec | ||
| + | ! scope=col | 45sec | ||
| + | |- | ||
| + | !temperature | ||
| + | |98° | ||
| + | |72° | ||
| + | |72° | ||
| + | |} | ||
| + | |||
| + | and last step: 72° during 10min | ||
| + | |||
| + | but we don't have any results | ||
| + | |||
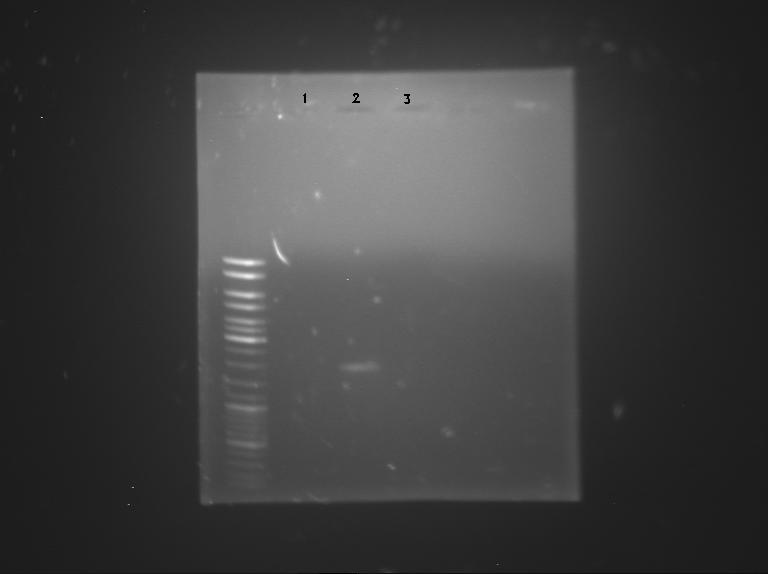
| + | ====Gel electrophoresis==== | ||
| + | [[File:Sean&Pierre&Melanie.JPG|400px|right]] | ||
| + | ''by Sean,Pierre & Melanie'' | ||
| + | |||
| + | Samples used: | ||
| + | # '''BBa_K762100''' (2µl) | ||
| + | # '''BBa_K517003''' (2µl) | ||
| + | # '''p cola'''(12µl) | ||
| + | |||
| + | ==Bibliography== | ||
| + | |||
| + | ''By Hoang Vu'' | ||
| + | |||
| + | Still working on my mission: find more informations about limonene, pinene and citral A and B.I got now a couple of very interessant reviews about limonene and pinene. | ||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 6_august.jpg|600px|center]] | ||
| + | |||
| + | '''Members present:''' | ||
| + | * Instructors and advisors: Alice. | ||
| + | * Students: Eugene, Fabio, Hoang Vu, Juliette, Melanie, Pierre, Romain, Sean and Terry. | ||
| + | |||
| + | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 14:21, 14 October 2014
Contents |
Wednesday 6th August
Lab work
The chassis coli Odor free
Final stock of MG1655
by Romain
2 final stocks of the MG1655 made with 1ml of strain and 250µl glycerol 87%, in -80°C.
Salicylate Inducible Suppressing System
Plasmid DNA Purification
by Eugene and Fabio
- BBa_K1372000 Cl.1
- BBa_K1372000 Cl.2
From Liquid Culture made the 5th August
Digestion
by Eugene and Fabio
In order to place the BBa_B0015 just after the BBa_K1372000, we will start the [http://parts.igem.org/Help:Assembly/3A_Assembly Assembly procedure] again.
- BioBrick BBa_K1372000 (Salicylate promoter + NahR + RNA suppressor) as Part A
- BioBrick BBa_B0015 (Double terminator) as Part B
The enzymes used were:
- EcoRI and SpeI for BBa_K1372000
- EcoRI and XbaI for BBa_B0015
TODO: Illustration of the process
BioBrick Assembly - Digest Reaction Protocol
Segregate Process by Electrophoresis
by Eugene and Fabio
- BBa_B0015 digested by EcoRI and XbaI
- BBa_K1372000 digested by EcoRI and SpeI
BioBrick Assembly - Segregate Process Protocol
Lemon scent
PCR Clean-up
by Pierre & Sean
BioBricks used: BBa_K762100 and BBa_K517003 (PCR perfomed on the 4th August)
PCR digestion
by Pierre & Sean
Protocol
For each BioBrick (Bba_K517003 and BBa_K762100)mix the following into a microcentrifuge tube.
| component | volume |
|---|---|
| H2O | 5.5μl |
| PacI | .5μl |
| Fast Digest buffer | 4μl |
| Cleaned-up PCR content | 30μl |
Then bain-marie at 37°C for one hour. Finally conduct PCR cleanup for both BioBricks, with 15µl of NE buffer.
We acidentally used PvuI instead of PacI. - SC 6/8/14
PCR pCola plasmide (Geranyl Synthase)
by melanie
amplification of geranyl Synthase
PCR protocol: 98° --> 2min
5 PCR cycle:
| time | 10sec | 20sec | 45sec |
|---|---|---|---|
| temperature | 98° | (depending on the tube - see the gradient) | 72° |
25 PCR cycle
| time | 10sec | 20sec | 45sec |
|---|---|---|---|
| temperature | 98° | 72° | 72° |
and last step: 72° during 10min
but we don't have any results
Gel electrophoresis
by Sean,Pierre & Melanie
Samples used:
- BBa_K762100 (2µl)
- BBa_K517003 (2µl)
- p cola(12µl)
Bibliography
By Hoang Vu
Still working on my mission: find more informations about limonene, pinene and citral A and B.I got now a couple of very interessant reviews about limonene and pinene.
Photo of the Day
Members present:
- Instructors and advisors: Alice.
- Students: Eugene, Fabio, Hoang Vu, Juliette, Melanie, Pierre, Romain, Sean and Terry.
 "
"