Team:Paris Saclay/Notebook/August/26
From 2014.igem.org
(→D - Lemon Scent) |
m (→Photo of the Day) |
||
| (3 intermediate revisions not shown) | |||
| Line 62: | Line 62: | ||
|} | |} | ||
| + | |||
| + | [[File:260814 verif chromo.jpg|400px|right]] | ||
D : sample digested by XbaI and PstI | D : sample digested by XbaI and PstI | ||
| Line 67: | Line 69: | ||
D1 - D2 - D3 - D4 - D5 - D6 - T - L | D1 - D2 - D3 - D4 - D5 - D6 - T - L | ||
| - | |||
We can see two or three bands for the digested samples : the digestion has worked. | We can see two or three bands for the digested samples : the digestion has worked. | ||
| Line 164: | Line 165: | ||
|8μl | |8μl | ||
|} | |} | ||
| + | |||
| + | |||
| + | [[File:260814 verif LS dans pGEMTeasy.jpg|300px|left]] | ||
D : sample digested by SacI | D : sample digested by SacI | ||
| Line 170: | Line 174: | ||
L - TLS2 - DLS2 - TLS3 - DLS3 | L - TLS2 - DLS2 - TLS3 - DLS3 | ||
| - | |||
The results are weird, the bands on the gel don't match our expectations, we don't know what happened with this plasmid. We should see two bands because the limonene synthase gene were inside the plasmid but we only can see one band which doesn't match with the size of pGEMTeasy+LS or with pGEMTeasy empty... | The results are weird, the bands on the gel don't match our expectations, we don't know what happened with this plasmid. We should see two bands because the limonene synthase gene were inside the plasmid but we only can see one band which doesn't match with the size of pGEMTeasy+LS or with pGEMTeasy empty... | ||
| Line 179: | Line 182: | ||
[https://2014.igem.org/wiki/index.php?title=Team:Paris_Saclay/Notebook/August/25#Transformation_of_DH5a_with_the_ligation Yesterday] we have transformed DH5A with pPS5 (PPSI +CAD. So we do a culture with some clone that have growth on the petri dish | [https://2014.igem.org/wiki/index.php?title=Team:Paris_Saclay/Notebook/August/25#Transformation_of_DH5a_with_the_ligation Yesterday] we have transformed DH5A with pPS5 (PPSI +CAD. So we do a culture with some clone that have growth on the petri dish | ||
| - | === | + | ===Lemon scent=== |
==== Extraction of pPS3 and pPS4==== | ==== Extraction of pPS3 and pPS4==== | ||
''by Terry'' | ''by Terry'' | ||
| Line 188: | Line 191: | ||
Stocks of this clones have been made. | Stocks of this clones have been made. | ||
| + | |||
| + | |||
| + | ==Photo of the Day== | ||
| + | [[File:Paris Saclay 26_august.jpg|400px|center]] | ||
| + | |||
| + | '''Members present:''' | ||
| + | * Instructors and advisors: Alice, Solenne and Sylvie. | ||
| + | * Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry. | ||
| + | |||
{{Team:Paris_Saclay/notebook_footer}} | {{Team:Paris_Saclay/notebook_footer}} | ||
Latest revision as of 15:28, 14 October 2014
Tuesday 26th August
Lab work
Construction of the fusion protein
plasmid extraction : pGEMTeasy + chromoprotein
by Laetitia
We used the bacteria containing pGEMTeasy + chromoprotein launched by Laetitia monday the 25th for the plasmid extraction, 6 independent cultures which come from the 6 stocks made yesterday.
To extract the plasmid, we used the plasmid DNA purification kit (Macherey-Nagel). Protocol for low copy plasmid.
Digestion of pGEMTeasy+chromoproteinby XbaI and PstI
by Laetitia
The goal is to check the presence of the insert inside the plasmid. We digest the 6 samples of purified plasmid.
| component | volume |
|---|---|
| Plasmid | 10 μl |
| Fast Digest buffer 10X | 1,5 μl |
| XbaI | 0.5μl |
| PstI | 0.5μl |
| H20 | 2,5 μl |
3-4h at 37°C
Electrophoresis of the digestion product of pGEMTeasy+chromoprotein
by Laetitia
T : control negatif without enzyme
| component | volume |
|---|---|
| blue | 2 μl |
| Plasmid | 2 μl |
| H20 | 8μl |
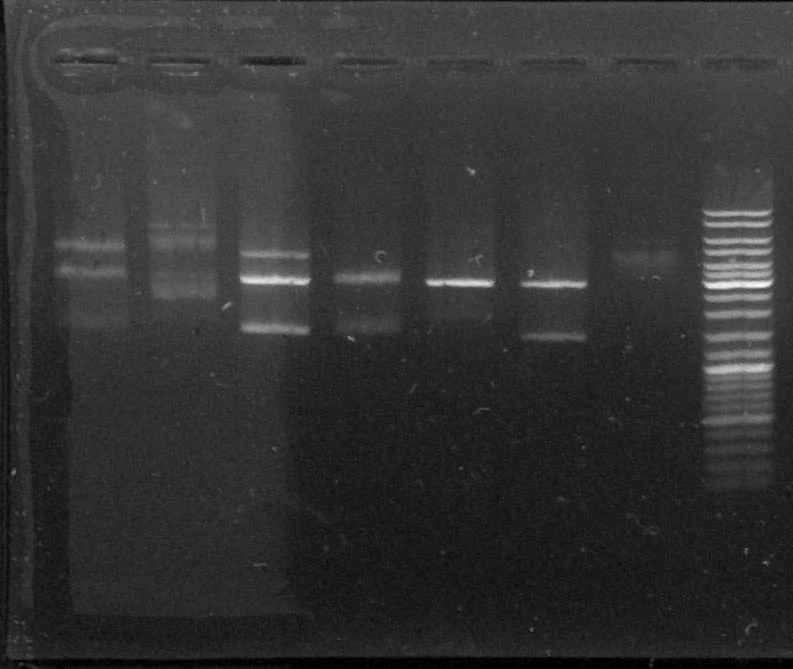
D : sample digested by XbaI and PstI
D1 - D2 - D3 - D4 - D5 - D6 - T - L
We can see two or three bands for the digested samples : the digestion has worked.
The band at 1500kb is the chromoprotein gene, the band at 3000kb is pGEMTeasy alone. The band at 4500kb is the plasmid containing the chromoprotein gene, non digested.
We decided to use the plasmid samples 3, 4 and 6
Digestion of pGEMTeasy+chromoprotein 3,4 and 6 by EcoRI and PstI
by Laetitia
We digest the remaining sample of plasmid extracted this day (20μl) : samples 3, 4 and 6
| component | volume |
|---|---|
| Plasmid | 20 μl |
| Fast Digest buffer 10X | 3 μl |
| EcoRI | 1μl |
| PstI | 1μl |
| H20 | 5 μl |
37°C - at night
Bacterial culture of pGEMTeasy+chromoprotein samples 3,4 and 6 from bacterial stock
by Laetitia
Culture in 5mL LB + ampiciline (1/1000) - 37°C at night
Lemon Scent
Streaking of colonies transformed by BBa_K762100+pGEMTeasy (LS)
Due to yesterday's strange results, we conduct 30 streakings in a petri dish: 28 white colonies and 2 blue colonies.
PCR of LS
by Mélanie
as previously described
Digestion of pGEMTeasy+ LS by SacI
by Laetitia
The goal is to check if the limonen synthase gene is really inside the plasmid. We used the two samples named " LS2 " and " LS3 " which are normally pGEMTeasy+LS .
| component | volume |
|---|---|
| Plasmid | 20 μl |
| Fast Digest buffer 10X | 3μl |
| SacI | 1μl |
| H20 | 6μl |
Electrophoresis of the digestion products by SacI
by Laetitia
T : control negatif without enzyme
| component | volume |
|---|---|
| blue | 2 μl |
| Plasmid | 2 μl |
| H20 | 8μl |
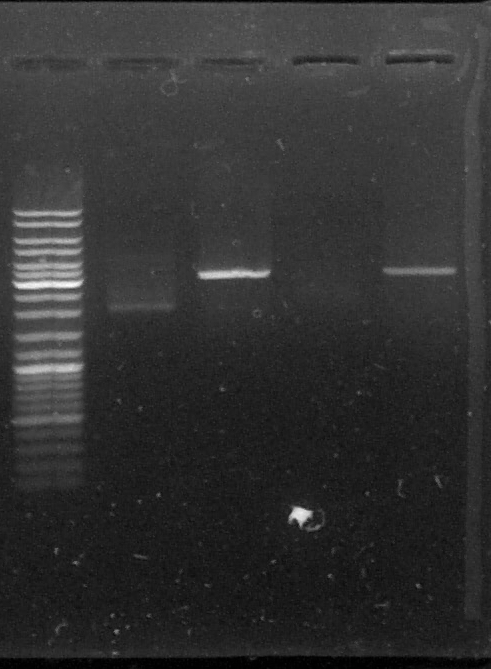
D : sample digested by SacI
L - TLS2 - DLS2 - TLS3 - DLS3
The results are weird, the bands on the gel don't match our expectations, we don't know what happened with this plasmid. We should see two bands because the limonene synthase gene were inside the plasmid but we only can see one band which doesn't match with the size of pGEMTeasy+LS or with pGEMTeasy empty...
Culture of bacteria with pps5
by Mélanie
Yesterday we have transformed DH5A with pPS5 (PPSI +CAD. So we do a culture with some clone that have growth on the petri dish
Lemon scent
Extraction of pPS3 and pPS4
by Terry
Extraction of plasmid pPS3 (coding beta-pinene) and pPS4 (coding geraniol) with the following protocol
2 different clones with pPS3 and 4 with pPS4 have been used for this extraction.
Stocks of this clones have been made.
Photo of the Day
Members present:
- Instructors and advisors: Alice, Solenne and Sylvie.
- Students: Eugène, Hoang Vu, Laëtitia, Mélanie, Romain, Sean and Terry.
 "
"