Team:Heidelberg/pages/Collaborations
From 2014.igem.org
(→iGEM Team Imperial Collage London) |
|||
| Line 3: | Line 3: | ||
=iGEM Team Aachen= | =iGEM Team Aachen= | ||
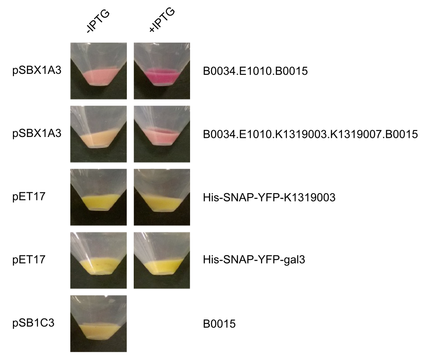
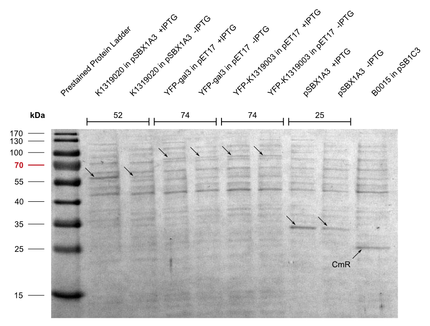
| - | Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, in exchange they offered to | + | Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, which they used to express their part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1319003 E1010-K1319003], the human galectin-3 protein fused to mRFP. Nicely seen in Figure 1 and 2 the the intensity of the red color is significantly higher for the induced sample with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362091 pSBX1A30] compared to the uninduced ones. To ensure the right molecular mass of the fusion protein, a SDS gel was run as well (Figure 3). |
| + | {{:Team:Heidelberg/templates/image-quarter| align=right|descr=| | ||
| + | caption=Expression of Galectin-3| file=AachenCo1.png}} | ||
| + | {{:Team:Heidelberg/templates/image-quarter| align=right|descr=| | ||
| + | caption=Expression of Galectin-3| file=AachenCo2.png}} | ||
| + | {{:Team:Heidelberg/templates/image-quarter| align=right|descr=| | ||
| + | caption=SDS-PAGE of K1319020 expression| file=AachenCo3.png}} | ||
| + | |||
| + | More information about their project can be found on Aachens [https://2014.igem.org/Team:Aachen/Project/Gal3 Galectin-3 site]. | ||
| + | In exchange they offered to characterised these expression vectors with their own designed analyser by measuring the amount of produced fluorescent proteins. Unfortunately, transporting <i>E.colis</i> to another city wasn't as easy as expected, not all Bacteria were able to grow afterwards and we could only obtain sufficient data from pSBX4C5. | ||
=iGEM Team Tuebingen= | =iGEM Team Tuebingen= | ||
Revision as of 20:34, 17 October 2014
Kurze Einleitung
Contents |
iGEM Team Aachen
Michael from the iGEM team Aachen visited us in Heidelberg. Their team worked on the development of a real-time pathogen detection technique for what they have build their own fluorescence measurement camera. We provided our expression vectors to the iGEM team Aachen, which they used to express their part [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1319003 E1010-K1319003], the human galectin-3 protein fused to mRFP. Nicely seen in Figure 1 and 2 the the intensity of the red color is significantly higher for the induced sample with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1362091 pSBX1A30] compared to the uninduced ones. To ensure the right molecular mass of the fusion protein, a SDS gel was run as well (Figure 3).
More information about their project can be found on Aachens Galectin-3 site. In exchange they offered to characterised these expression vectors with their own designed analyser by measuring the amount of produced fluorescent proteins. Unfortunately, transporting E.colis to another city wasn't as easy as expected, not all Bacteria were able to grow afterwards and we could only obtain sufficient data from pSBX4C5.
iGEM Team Tuebingen
iGEM Team Freiburg
Three of our team members, Anna, Caro, and Charlotte visited the iGEM team Freiburg. We met the team right after a big team meeting, therefore we were able to hear their latest project developments. We spend the evening talking about experiences in the lab and about iGEM. It was relieving to hear that they faced similar difficulties in the lab. The morning after we presented our project to the iGEM team Freiburg. Freiburg was very interested in our program iGEM@home as we offered them to promote their own human practise projects. Both our teams are working with light inducible systems, manly the LOV domain. Although working in different organisms, the constructs were compatible, therefore we could interchange protocols for induction and preliminary results. Furthermore we gazed at their professional light induction box - we can definitely improved ours.
Gibson vs. Cpec?
After cooperation with the iGEM Team Freiburg for two years in a row we are hoping to keep Freiburg as a reliable partner in the next years as well.
 "
"