Team:ETH Zurich/expresults/rr
From 2014.igem.org
Riboregulators
As many former iGEM teams have encountered unwanted basal expression of their genes of interest (goi) due to promoter leakiness (ETH2013, Groningen2012 amongst others), we investigated this severe obstacle to tightly controllable systems. We looked into the available literature and found with riboregulators a promising approach to challenge the issue of leakiness[32]. In addition, we chose this approach, to study and characterize such regulators combined with quorum sensing modules and make the resulting parts available for the iGEM community in the [http://parts.igem.org/Main_Page Registry of Standard Biological Parts].
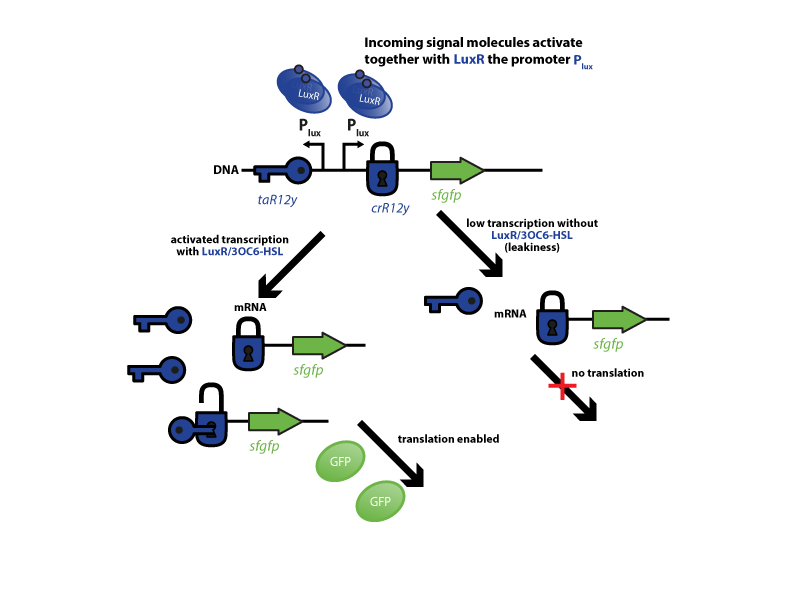
The riboregulator systems include two parts: 1) a cis-repressed RBS in front of the goi, and 2) a co-expressed trans-activating RNA.
To characterize this system for reliable cell-to-cell communication, we combined the riboregulator part with promoters of the quorum-input-sensing systems used by our team (LuxI/LuxR, LasI/LasR, and RhlI/RhlR). In our final gene circuit, the riboregulators were intended for the control of integrases. However, to have an easily quantifiable output, we used GFP as our riboregulated goi and measured the output fluorescence with a Tecan plate reader. This approach is described below in the figure 1.
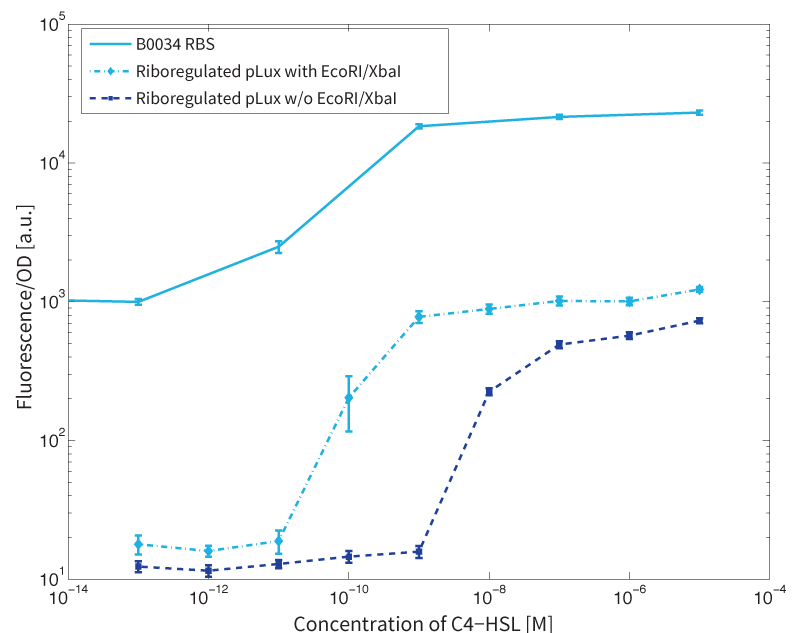
First, the experiments were conducted with cells transformed with two different pPaB plasmids. These plasmids contained either the pBR322 origin (pMB1) or the p15A origin, which yielded a stable two-plasmid system with about 15-20 copies per plasmid and cell (medium-copy). Also, exactly the published riboregulator sequences were used in the the first place to have a positive control for functionality. This is important, because the original riboregulator sequence includes two forbidden restriction sites (EcoRI and XbaI). Second, the restriction sites were removed by two different approaches: a) multiple site-directed mutagenesis, b) blunting and ligation (Klenow and T4 DNA polymerase). Afterwards, the functionality of the construct was again tested to approve that no severe loss-of-function occurred due to the site-removal. In a third step, the construct was transferred into the [http://parts.igem.org/Part:pSB1C3 pSB1C3] backbone (pMB1 origin, high copy number of 100-300) to be in line with the [http://parts.igem.org/Help:Standards/Assembly BioBrick Standard]. Below, you can find the corresponding graphs in fig. 2 and fig. 3, respectively.
In more detail, we have characterized promoters of the three quorum-sensing systems. These are pLux, pLas, and pRhl. To have a reference, we first measured the transfer functions with a non-regulated RBS in front of sfGFP, i.e. the amount of fluorescence in dependence of the inducer concentration present. Next, we repeated the experiments with the corresponding riboregulated constructs once with and without the forbidden restriction sites.
To conclude, we found an about 60-fold reduced basal GFP expression and an increased signal-to-noise ratio of about 6-fold when using our riboregulator construct, as compared to a non-regulated [http://parts.igem.org/Part:BBa_B0034 reference RBS (B0034)]. These results are summarized in figure 2. Finally, the BioBrick conform module was confirmed to show no loss-of-funtion due to EcoRI and XbaI site removal. However, the sensitivity towards the inducer was reduced (see figure 3).
 "
"