Team:ETH Zurich/blog
From 2014.igem.org
{{{1}}}
Blog
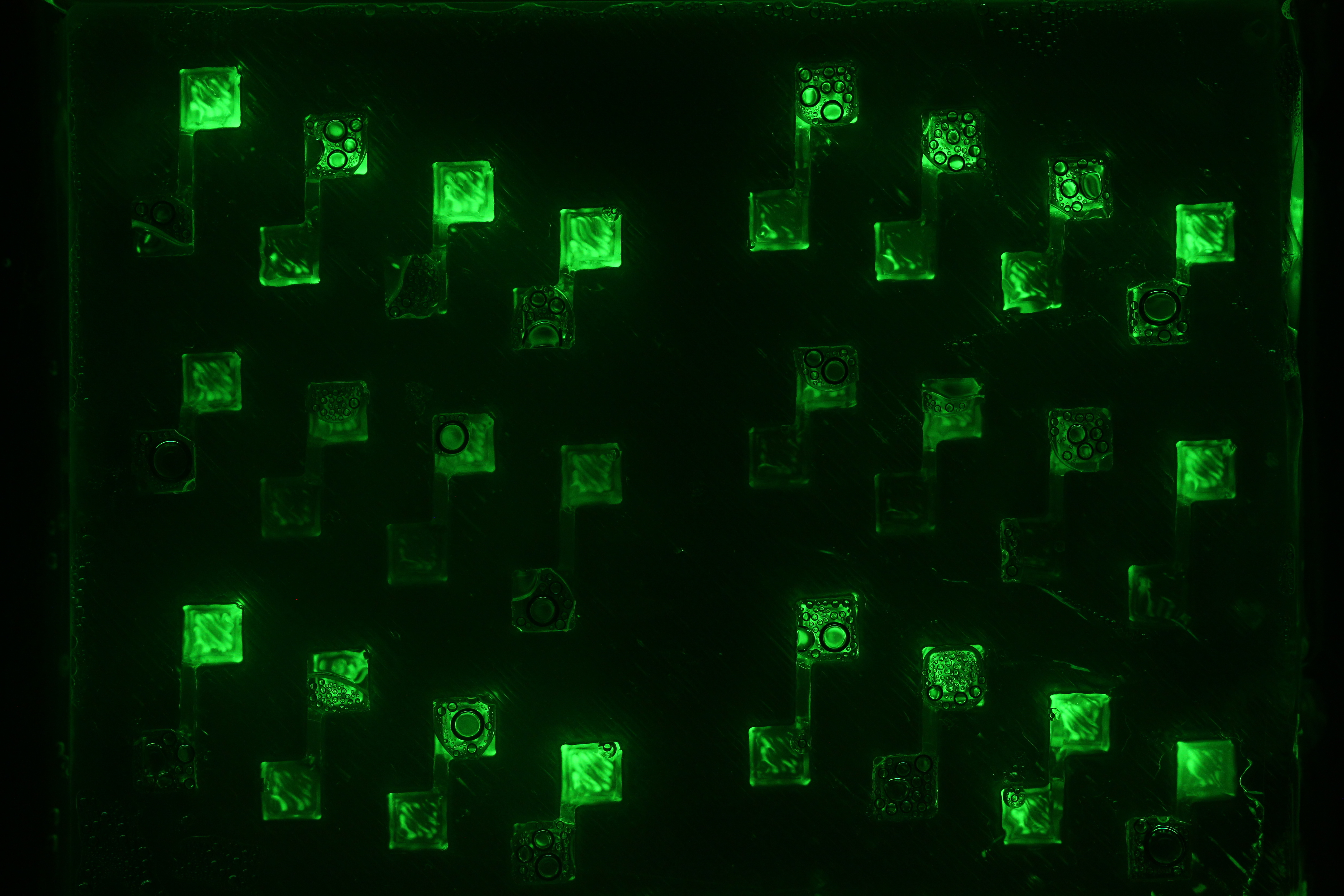
Diffusion Experiments
Monday, 25th August
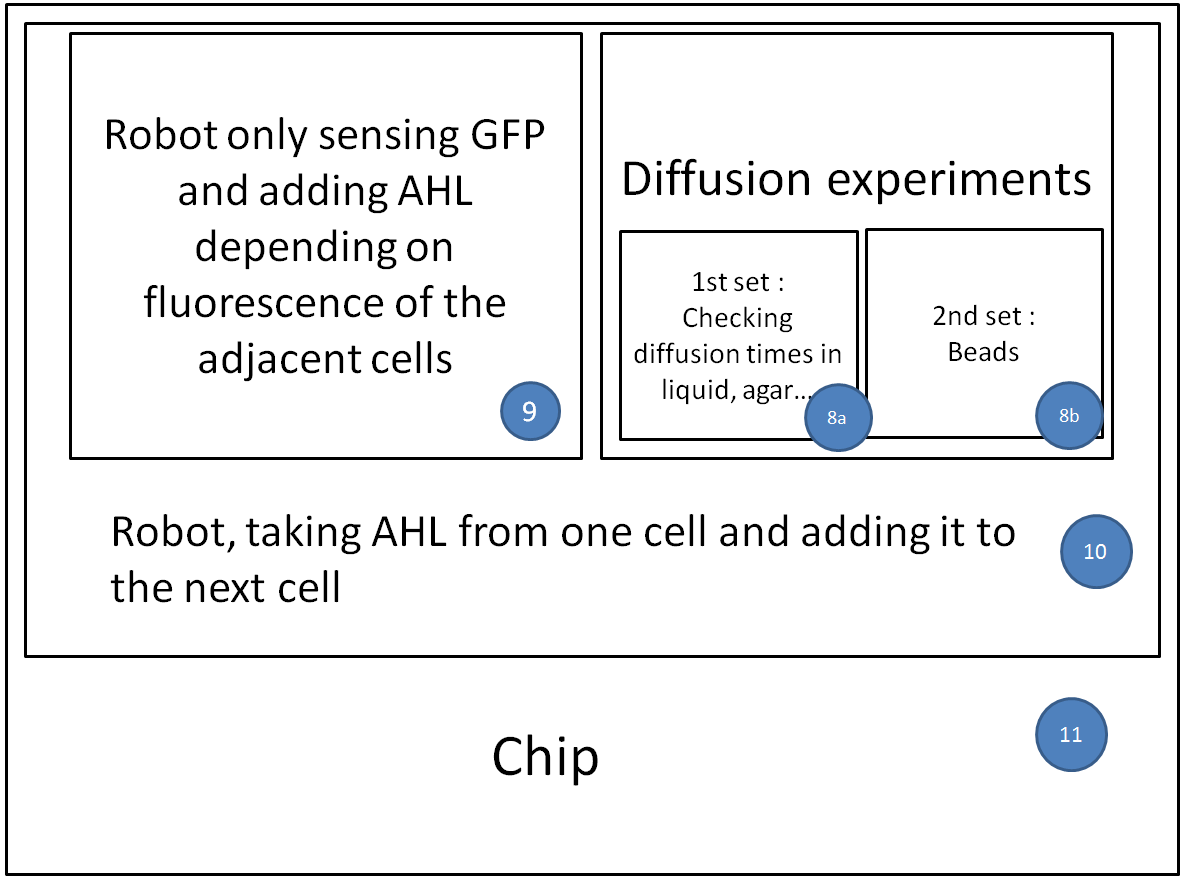
Now that we tested the different constructs for leakiness, dose response and cross talk, we are investigating in the diffusion rate of AHL. Therefore we designed a chip with chambers connected by channels of different length (see figure). In one chamber we added a strain containing a regulator and a sensor plasmid, to the second chamber we added AHL or an AHL producer strain. As soon as AHL is diffused to the former chamber it induces sfGFP production on the sensor plasmid. The time it takes until we can measure the sfGFP signal and the length of the channel gives us information about the velocity of AHL diffusion.
Three main experiment designs were tested and modified: the agar-design, the liquid-design and the beads-design.
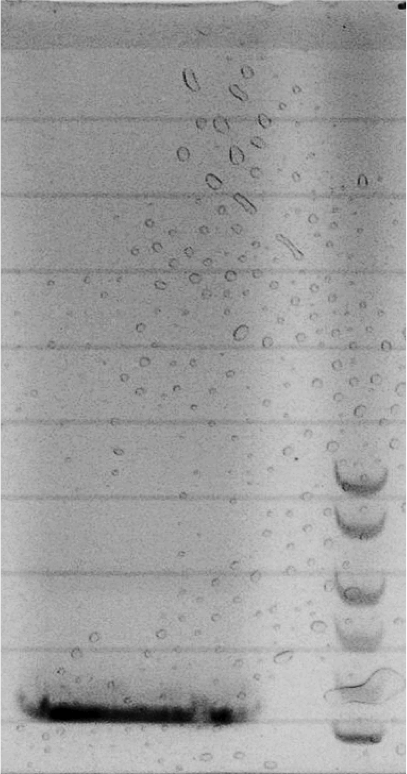
- For the agar-design we filled the chamber and channels with LB-agar or in a second attempt with agarose only. Subsequently, we punched a cylindric hole in each chamber. The capacities were filled with regulator/sensor strain culture or AHL/AHL producer strain culture, respectively. After 3 to 4 h at 37 °C we could observe an increase in the sfGFP signal. Since we used comparably high concentration of AHL in our first attempts, we could not detect a delayed sfGFP signal for chambers connected by a longer channel. The experiment will be repeated using lower concentrations. However, conducting the experiment using the agar-design, we encountered a problem: after some hours the LB-agar/agarose started to shrink and dried out. Thus, the agar-design cannot be used without modifications for experiments of longer duration.
- In an attempt we only left a small agar fragment in each channel and filled the rest with liquid medium, so as to decrease chance of it drying out, while avoiding at the same time that bacteria from one chamber reach the other chamber.
- The bead-design consists of an alginate bead for each chamber and liquid medium in between. The beads either contained regulator/sensor-bacteria or AHL.

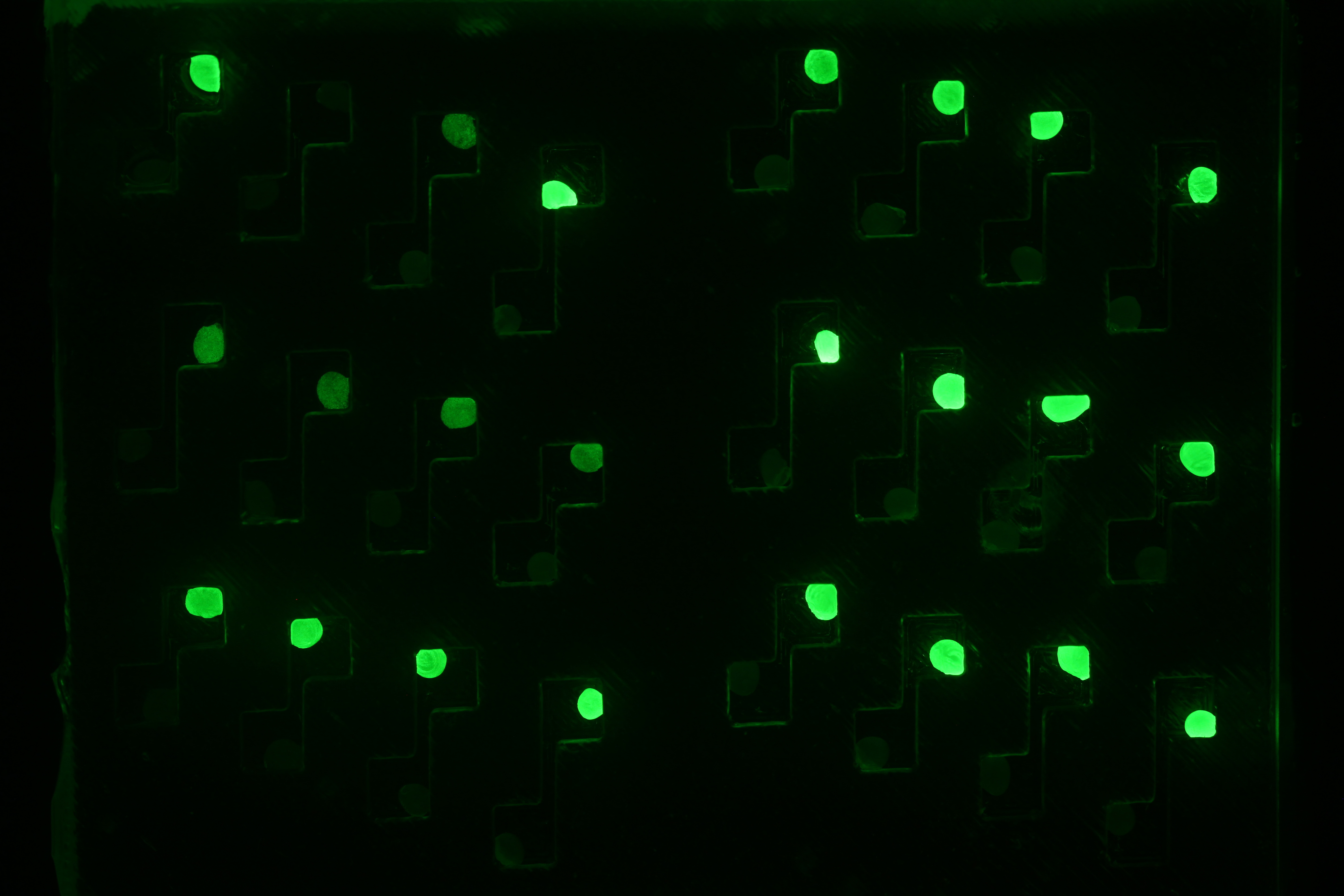
First cross talk experiments
Friday, 11th August
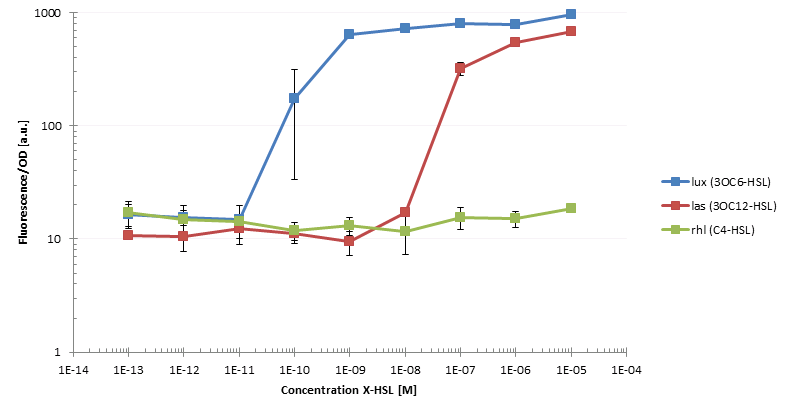
After having constructed the necessary regulator and sensor plasmids we were able to start first cross talk experiments. Therefore we transformed competent cells with different combinations of regulator and sensor plasmids. One example would be the strain siG0024 which contains the regulator plasmid piG0041 producing LuxR under a constitutive promoter and the sensor plasmid piG0051 that encodes for sfGFP. The latter is under control of a pluxR promoter, hence it will be transcribed in the presence of LuxR and Lux. To assess crosstalk and leakiness we conducted multiple experiments: after adding one of the three AHLs to the cultures we analyzed the raise in sfGFP production using a microtiterplate reader. In a first step we measured the dose response of constructs with different ribosomal binding sites (RBSs) and with or without riboregulator. We could observe differences not only in sensitivity, but also in leakiness; some strains expressed GFP even without AHL induction. To give an example: we evaluated the sensitivity of the siG0024 strain to different concentrations of 3OC6-HSL (LuxI product) and compared its characteristics to those of other constructs. Additionally, we measured the sensitivity of siG0024 to 3OC12-HSL (LasI product) and C4-HSL (RhlI product). Indeed, we could observe the induction of sfGFP transcription by comparably low levels of 3OC12-HSL. Crosstalk can be observed between the Las and the Lux system. To simulate a situation most similar to the final experiment, we added the diluted supernatant of AHL producer cells to the test cultures. However, we could not detect any GFP production as reaction to the supernatant. We assume that the AHL concentration is not high enough to induce transcription. By constructing stronger RBSs on the producer plasmids we aim at increasing the AHL production. All these informations we will use to design a system tailored to our plans.
The figure shows the dose response of xxx to 3OC12-HSL, 3OC6-HSL and C4-HSL. Both 3OC12-HSL and 3OC6-HSL induce production of GFP, although at different concentrations. This is an example for cross talk between the lux and the las system.
- REDIRECT Team:ETH Zurich/labblog/20140829mod
Week 8 : Plasmdis assembly, First mold 3D printed
Wednesday, July 16th
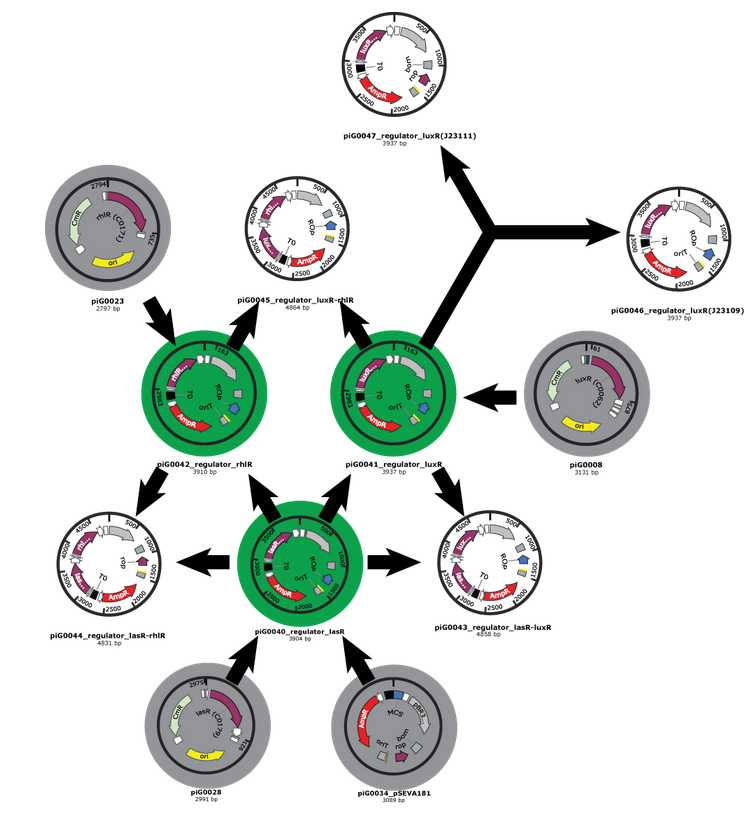
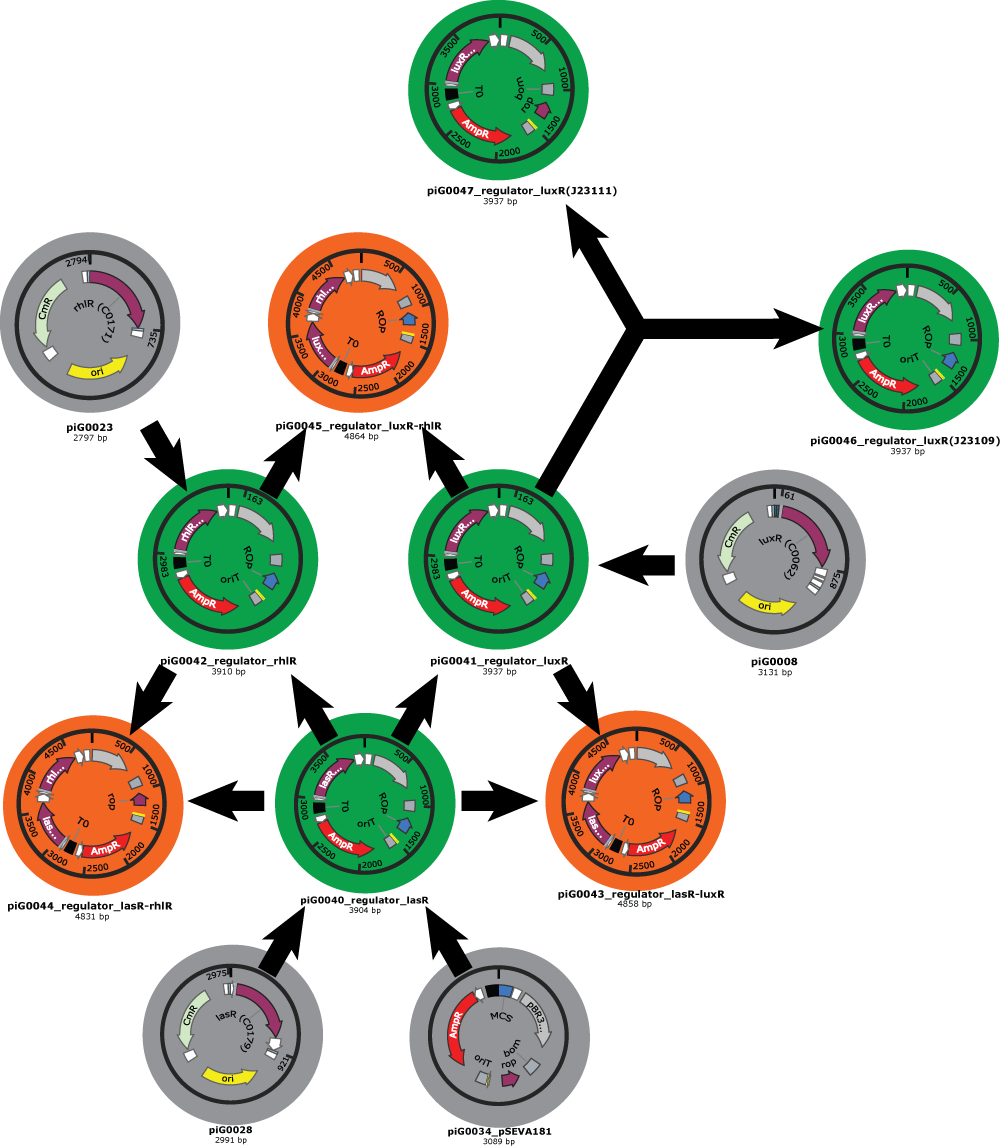
- Our first Gibson assemblies worked well. Regulator plasmids piG0040, piG0041 and piG0042 have been constructed :
- We printed our first mold for millifluidic chip :
- We simulated the whole model with and without riboswitch in Matlab with random parameters for integrases. We are still trying to estimate integrase parameters from the literature but it is very difficult.
Week 7: Human practice planning, Plasmid assembly running, Logo design
Wednesday, July 9th
- We have a logo !
- In the lab, we did some :
- Plasmid preparation
- Sequencing
- Digests
- Purification of backbone fragments needed for GA
- On the modeling side, we tried to estimate integrase parameters from the paper from Bonnet et al. [9], more particularly with the figure S4 :
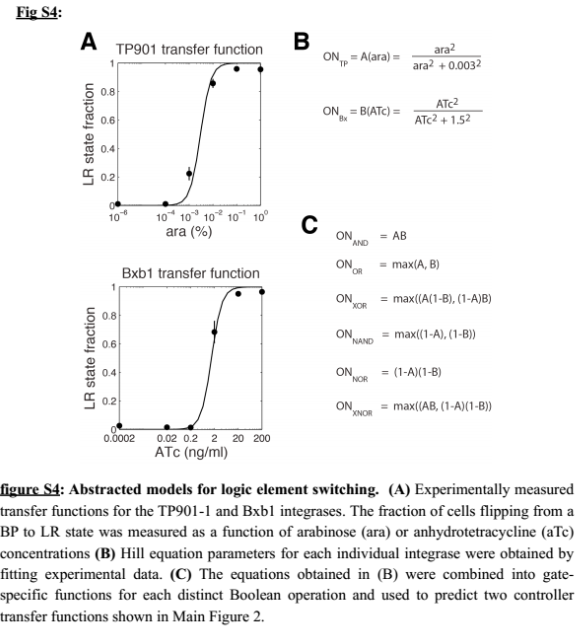
We are trying several strategies : minimization of the error function and Markov Chain Monte Carlo.
- We have a precise plan for our human practice project. We would like to study the emergence of complexity in many different fields and investigate how people deal with it. We will do this with interviews of experts in different fields, and with a survey for a wider outreach. We will also reach younger people by organizing talks in schools. We will finally give our own advice on the question by writing an essay on the subject and linking it to our experience with Mosaicoli.
Construction of the regulator plasmids
Thursday, July 5th
Construction of the lasR regulator plasmid (piG0040)
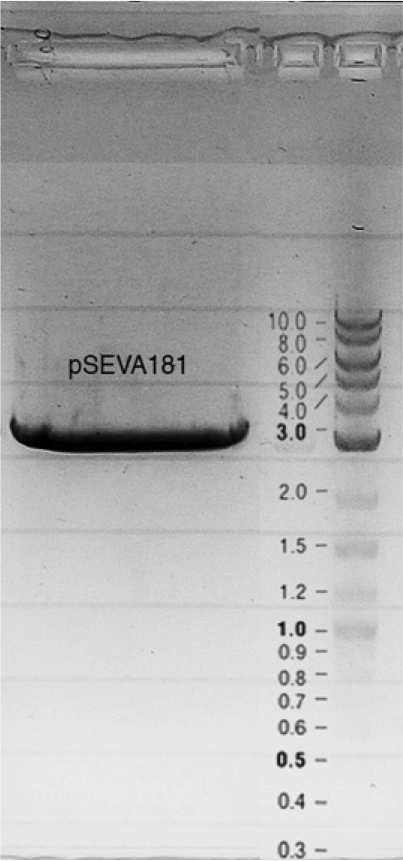
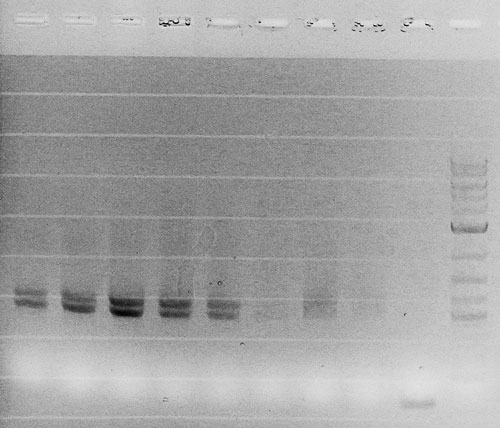
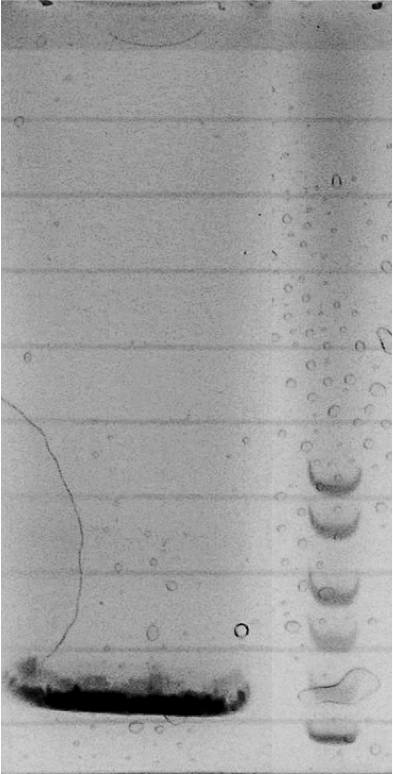
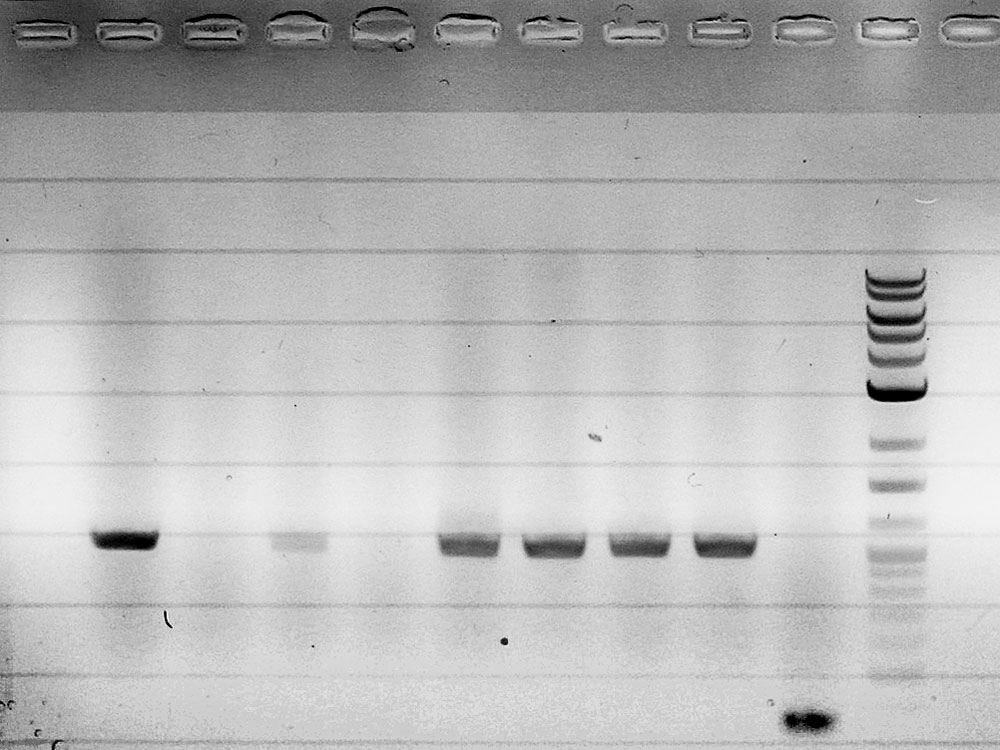
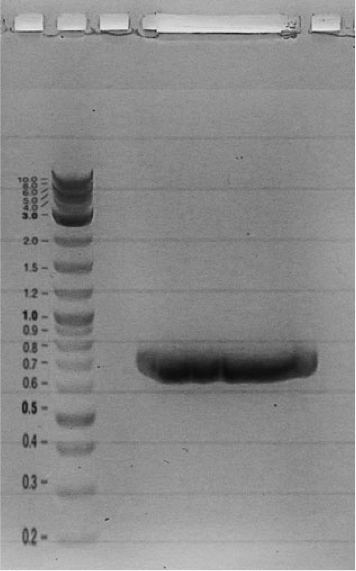
Competent cells were transformed with piG0028 (K553003, lasR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of lasR by PCR using oiG0003 and oiG0004 resulted in the fragment fiG0004 (1.0 kb). The vector piG0034 (pSEVA 181) was digested with the restriction enzymes HindIII and PacI to fiG0001 (3.0 kb). The two fragments, fiG0001 and fiG0004, were assembled using Gibbson assembly. Thus we used the plasmid backbone of piG0034 and lasR of piG0028 to construct the lasR regulator plasmid piG0040. Competent cells were transformed with piG0040 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.0 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the luxR regulator plasmid (piG0041)
Competent cells were transformed with piG0008 (F2620, luxR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of luxR by PCR using oiG0005 and oiG0006 resulted in the fragment fiG0005 (0.8 kb). The backbone of the vector piG0040 (lasR in pSEVA 181) was amplified by PCR using oiG0007 and oiG0008. This fragment, fiG0011 (3.2 kb), and fiG0005 were assembled using Gibbson assembly. Thus we formally replaced lasR by luxR to construct the luxR regulator plasmid piG0041. Competent cells were transformed with piG0041 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the rhlR regulator plasmid (piG0042)
Competent cells were transformed with piG0023 (C0171, rhlR) and selected on chloramphenicol-LB-plates. Plasmid DNA was extracted by performing a miniprep. The relevant plasmid sequence was sequenced by Microsynth using the primers oiG0001 and oiG0002. Amplification of rhlR by PCR using oiG0009 and oiG0010 resulted in the fragment fiG0006 (0.8 kb). The backbone of the vector piG0040 (lasR in pSEVA 181) was amplified by PCR using oiG0011 and oiG0012. This fragment, fiG0012 (3.2 kb), and fiG0006 were assembled using Gibbson assembly. Thus we formally replaced lasR by rhlR to construct the rhlR regulator plasmid piG0042. Competent cells were transformed with piG0042 and selected on ampicillin-LB-plates. Colony PCR using the primers oiG0035 and oiG0036 was conducted to check the size of the inserted fragment (expected bands at 1.1 and 1.2 kb). The sequence was verified by Microsynth using the same primers.
Construction of the luxR regulator plasmids with alternative constitutive promoters (piG0046 and piG0047)
The promoter site of the luxR regulator plasmid piG0041 was mutated by QuikChange site-specific mutagenesis to produce promoters of different strength. The primers oiG0031 and oiG0032 were used to establish piG0047, a plasmid with a promoter of intermediate strength (J23111). Analoguos the primers oiG0033 and oiG0034 were used to construct piG0046, a plasmid with a weak promoter (J23109). Competent cells were transformed with piG0046 and piG0047 and selected on ampicillin-LB-plates. The sequence was verified by Microsynth using the primers oig0035 and oiG0036.
Week 6: Project planning, Gibson assemblies planning, First Matlab simulations
Wednesday, July 2nd
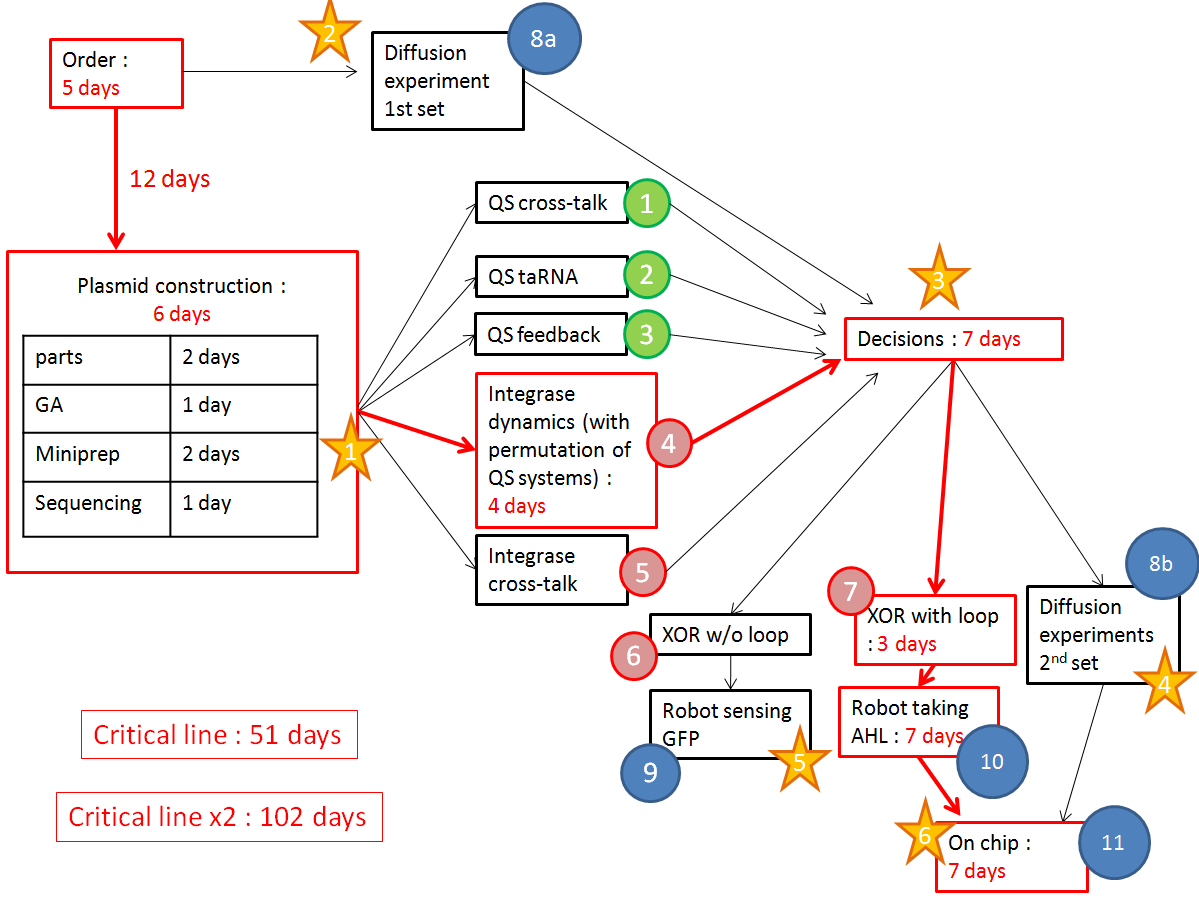
- We have a clear organization of the different experiments we would like to perform.
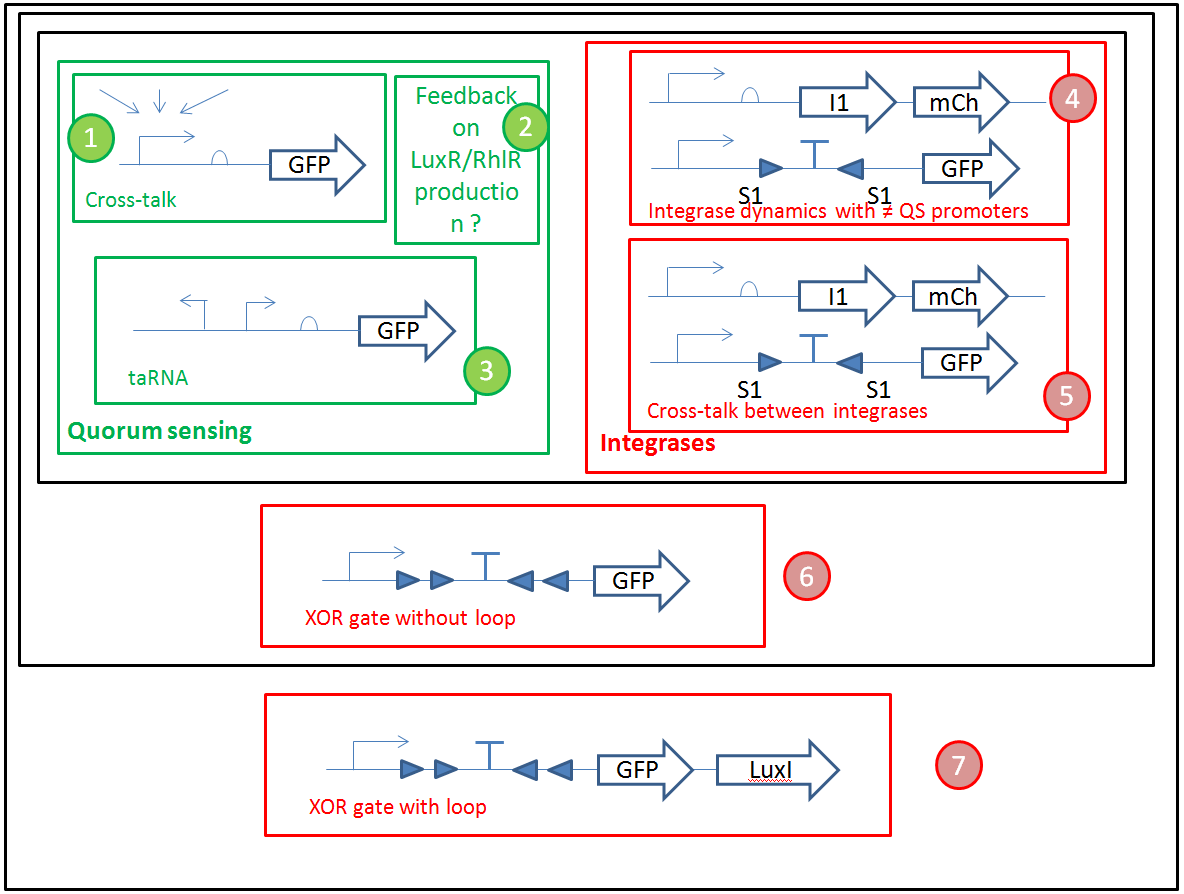
We want to optimize quorum sensing to have non-leaky non-cross-talking constructs. We want to prevent cross-talk between integrases and check their dynamics with different quorum sensing promoters. Then we will test the XOR gate without production of LuxI, to check how it works without the loop, and we will finally test our final construct with the loop, hoping that the delay between GFP production and the possible second switching due to AHL production is long enough to have enough fluorescence.
The red line is our critical timeline. Numbers of days are very optimistic,therefore we multiply the whole timeline by 2. The stars stand for modeling inputs.
- We have also clear plans for the Gibson assemblies to perform. As an example, here is the plan for regulator plasmids :
- We have a facebook page !
- We simulated quorum sensing without leakiness on Matlab, with parameters from the literature.
Week 5: Assembly strategy and steady states derivation
Wednesday, June 25th
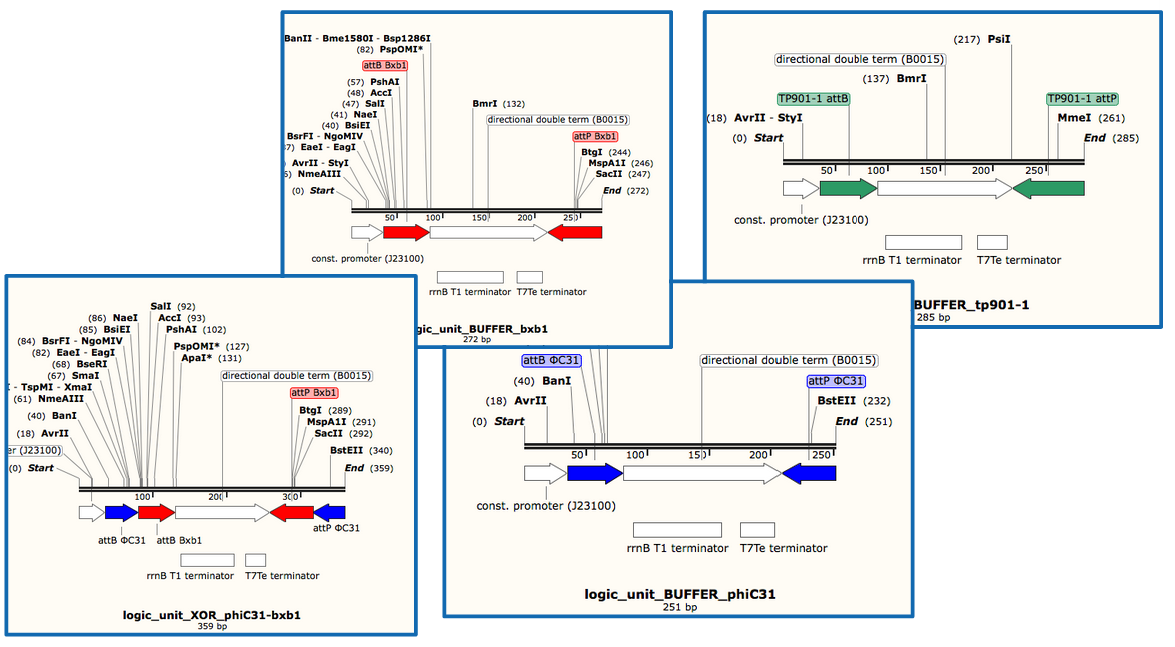
- We will use Gibson assembly to assemble our constructs, and have now a construct assembly strategy. We know the list of experiments we want to carry (cross-talk between quorum sensing molecules, quorum sensing leakiness with or without riboswitch, cross-talk between integrases, integrase activity and logic gate behaviour) and the exact sequences of base units we will assemble (lux, rhl, las promoter and repressor units, reporter units, logic units with 3 different integrases, constitutive AHL production units for diffusion test, backbone units).
- From the modeling side, we calculated analytically the steady state of the integrase and terminator modules, and it appears that the deterministic model gives a binary steady state corresponding to an XOR gate. This is what we expected, it confirms that deterministic model in our case is interesting for the study of dynamics of the whole system, and not for steady state calculations. So we are now moving to dynamic studies.
Week 4: Improving plasmid design, Modularization of the whole model
Wednesday, June 18th
- We improved a little bit the plasmid design, determined which parts will be taken from the iGEM registry, and which parts we have to synthesize.
Sensor plasmids will be approx. 9.3kb long, and logic plasmids will be approx. 5,2kb long.
- From the modeling side, the whole model has been modularized and we have calculated and simulated steady state for the quorum sensing module.
- GeneScript, Microsynth, EraSynbio are willing to support us !
Week 3: Plasmid design started, Modeling started
Wednesday, June 11th
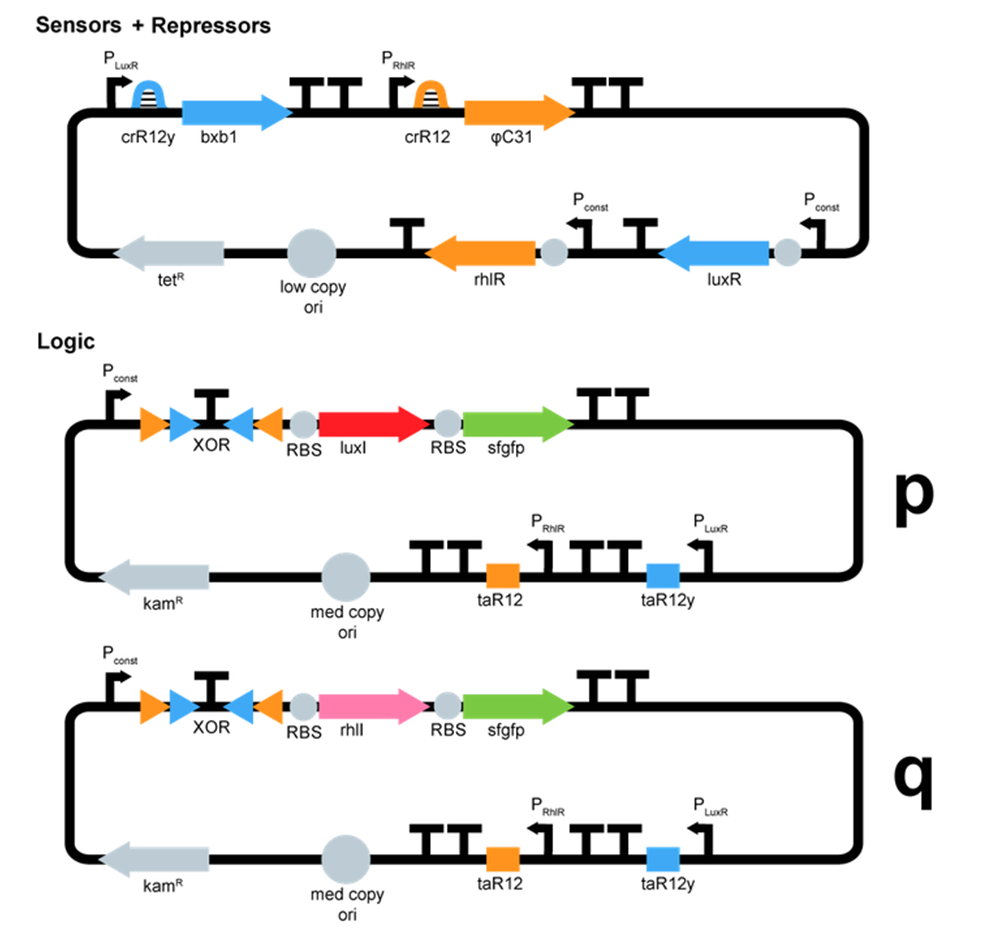
- We started plasmid design :
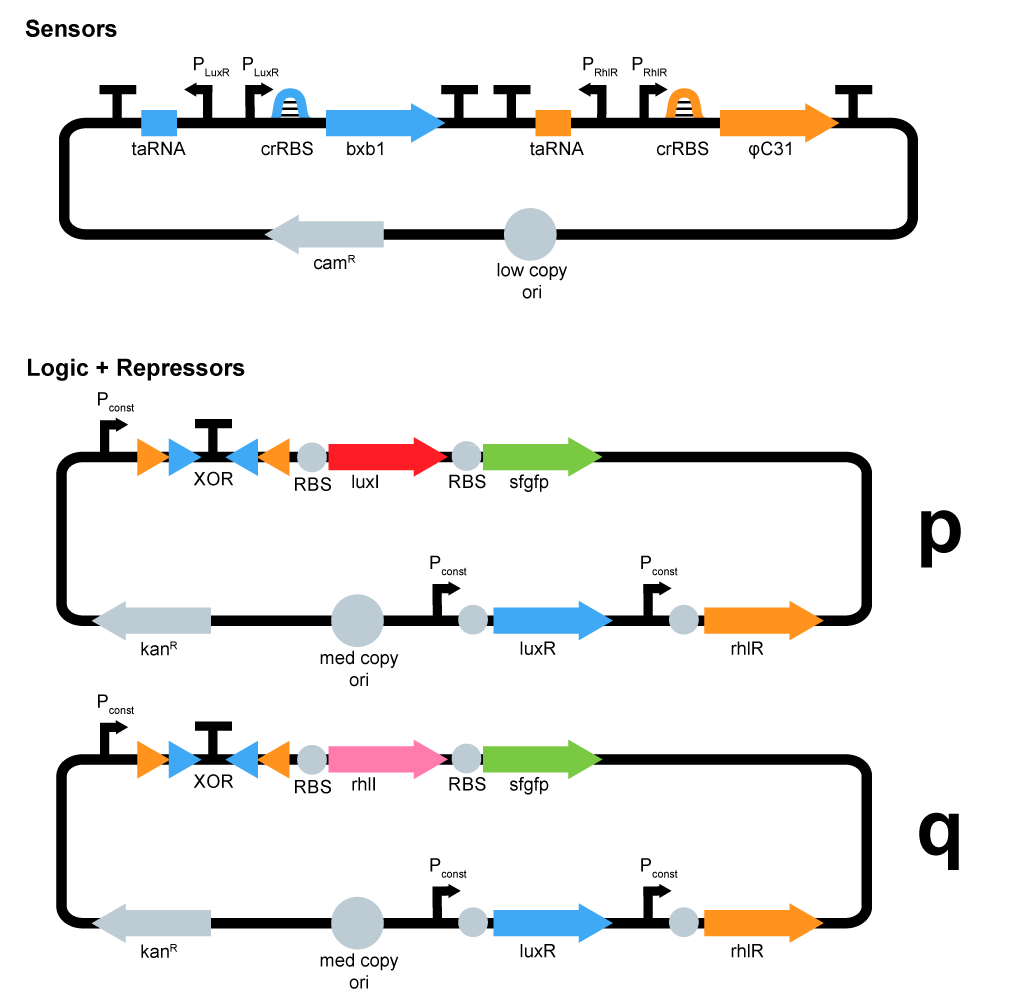
ΦC31 and Bxb1 are integrases. LuxR an RhlR are quorum sensing repressors. A riboswitch construct is placed around quorum sensing constructs to prevent leakiness of lux and rhl promoters. Type p colonies produce AHL by expressing the enzyme LuxI. Type q colonies produce Rhl by expressing the enzyme RhlI. In fact, we don't know yet which quorum sensing systems we will use. We will have to perform cross-talk experimetns in order to choose the ones that are the most orthogonal.
- We tried to print our first agar millifluidic chip : we printed it too small, and the printer had resolution problems.
- We wrote all reactions and found parameters from the literature for our model.
Week 2: Investigating microfluidics, Writing a modeling pipeline
Wednesday, June 4th
This week, we have talked with people from the microfluidics group and it came out that we should be able to use beads to encapsulate our cells and use a microfluidic chip. Therefore we will try to develop this possibility in parallel with the 3D-printed agar chip.
We have set up a modeling pipeline. We divided the modeling project into 3 parts :
- diffusion,
- parameter fitting,
- modeling of the genetic circuit itself, which comprises
- a deterministic model
- a stochastic model.
We know that the modeling challenges will be to model how integrases work, and to model the delay between reception of a quorum sensing signal and production of a quorum sensing signal by the receiver cell. Indeed, a colony could switch itself OFF when it should be ON, if it receives QS1 and produces QS2, for example. If a delay is present between reception of QS1 and production of QS2, the colony will produce GFP before it is switched OFF, and as GFP is stable enough, it will stay visible during a long time and the self-switching OFF won't be observed. This delay should be modeled.
Week 1 : Project selected
Wednesday, May 28th
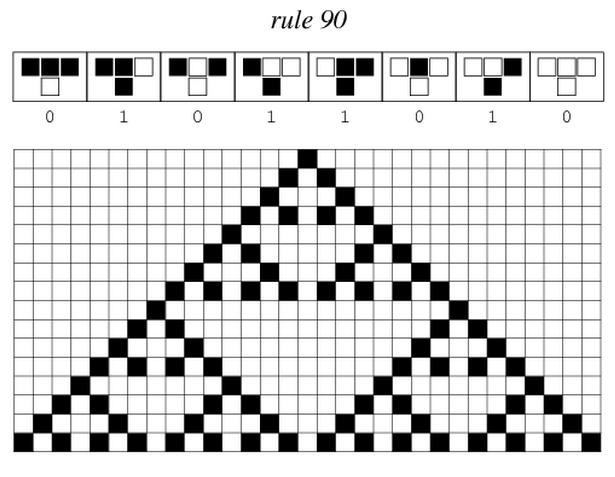
After more than one month of endless meetings and passionate debates, we finally chose the project that will keep us occupied in the next five months. From the beautiful pattern made by the Sierpinski triangles, we will focus on cellular automata and try to implement one.
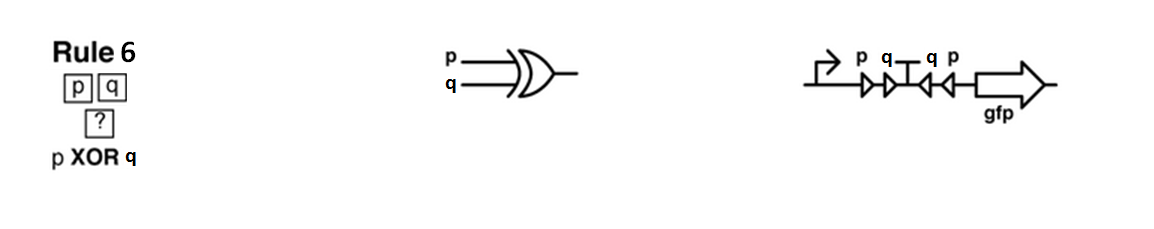
Sierpinski triangles appear when the rule 90 is followed by every cell on the grid :
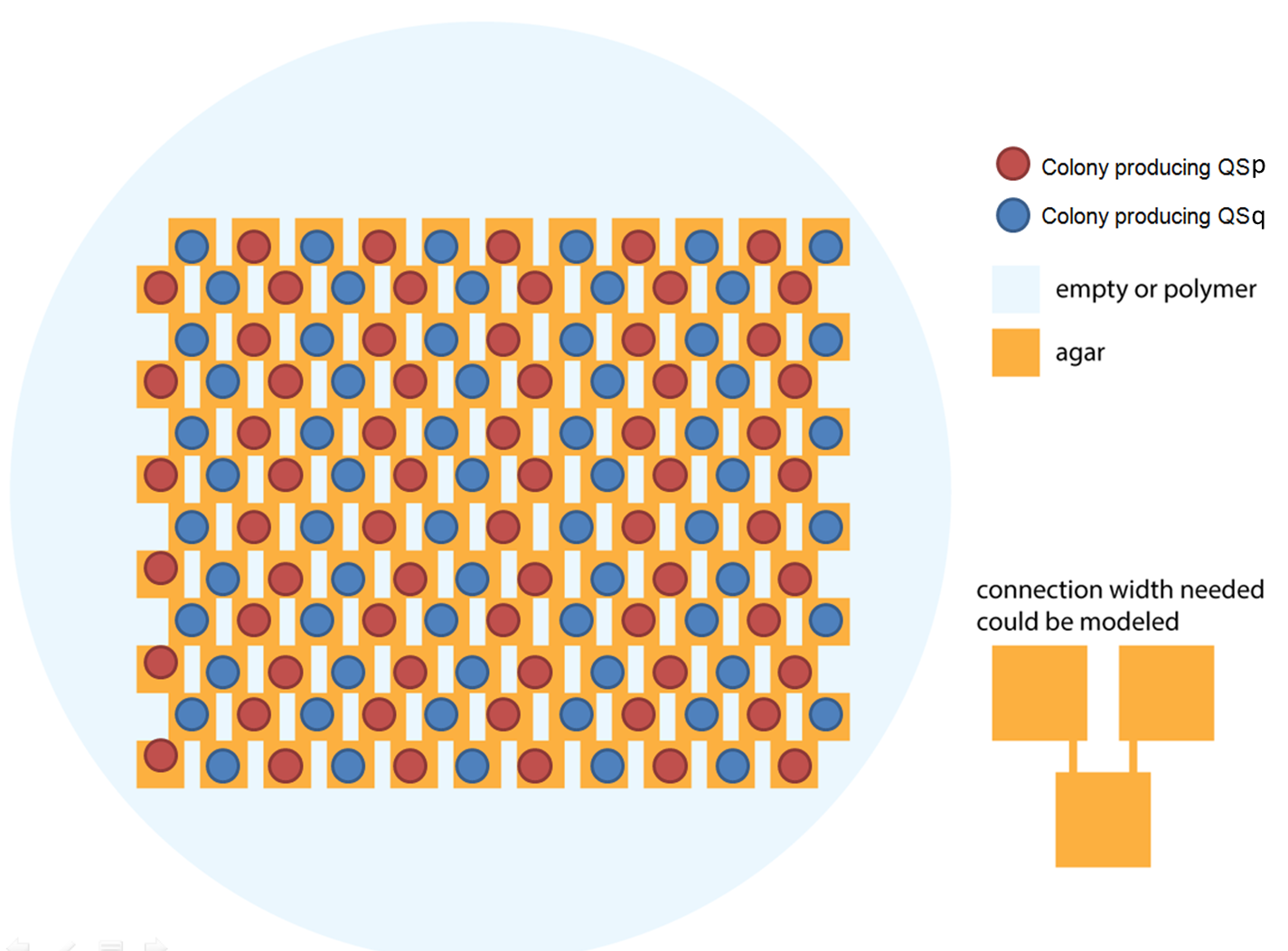
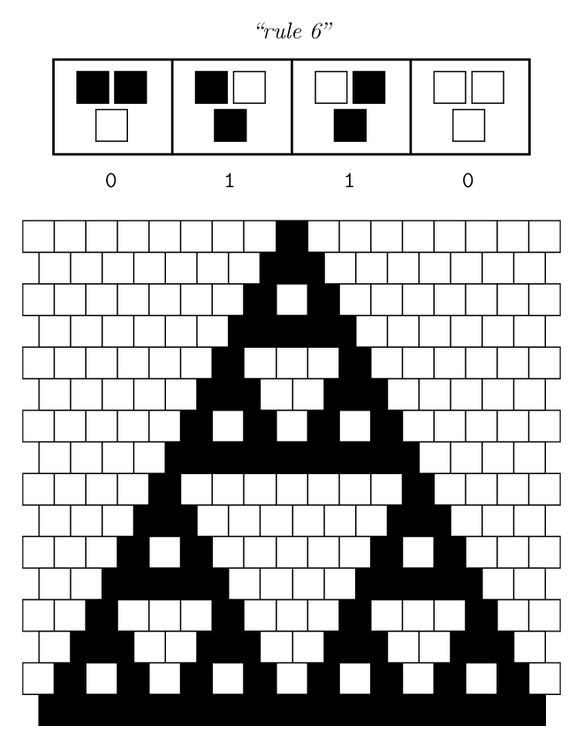
Ideally we will use a microfluidic chip. We could also use a 3D printed agar plate like this one to load the colonies. On this grid we can implement the rule 6, which is the simplification of rule 90 considered as a rule with 2 inputs : each cell computes a simple XOR gate of its two parents.
The logic part will be built with integrases and the colony-to-colony communication will use quorum sensing.
Every colony will receive two quorum sensing signals (QSp and QSq) from the two cells above it. These two signals trigger the production of two different integrases r and s in the colony. Integrases enable to build biological XOR logic gates by switching twice a terminator. Indeed, every integrase can switch the terminator only once. Thus if the colony produces only r or only s, the terminator is switched only once, so the terminator is OFF, and GFP and QS1 or QS2 are produced (depending on the colony). If the colony produces r and s, the terminator is switched twice, so it is ON and it blocks expression of GFP and of the quorum sensing molecule.
We need to :
- find orthogonal quorum sensing molecules and orthogonal integrases
- discuss with microfluidics experts to check if using microfluidics is possible and presents advantages in our case
- find possible parts in the registry for integrases, and design plasmids
BSSE Openhouse Day
Saturday, May 10th
Sharing our iGEM and synthetic biology interest with the public
On May 10th the public in Basel had the unique chance to get an insight into many different scientific laboratories and the work done there. It was the joint open house day of D-BSSE of ETH (Department of Biosystems Science and Engineering) and the Biozentrum of the University of Basel. The many different labs opened their doors to the public and many scientists were present to give interested people some details about their daily work. So did the ETH iGEM team 2014. The team was present with a poster showing the history of iGEM, the previous ETH iGEM teams with their projects and general information about synthetic biology. Additionally there was a slideshow giving a best-of photo collection of last years jamboree. The goal of this day was to inform the public about synthetic biology in general and specifically about the spirit and the many different projects of iGEM. Many people showed strong interest in truly student driven projects and are curious to follow our team wiki for the next months.
 "
"