Team:Heidelberg
From 2014.igem.org
(Difference between revisions)
| Line 250: | Line 250: | ||
</div> | </div> | ||
<div class="col-md-9 col-xs-12"> | <div class="col-md-9 col-xs-12"> | ||
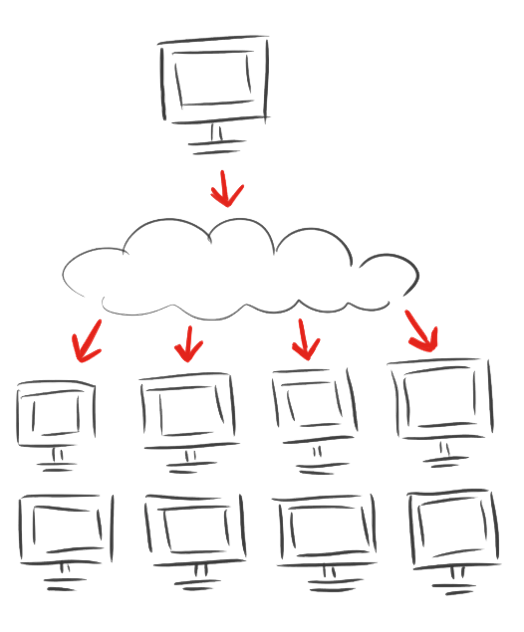
| - | <p class=" | + | <p class="align-right"><span class="larger">As calculations on protein structures are costly<br />we involved the rest of the world with</span></p> |
<h1 class="very-large-text red-text">iGEM@home</h1> | <h1 class="very-large-text red-text">iGEM@home</h1> | ||
</div> | </div> | ||
| Line 257: | Line 257: | ||
<div class="col-md-8 col-xs-12"> | <div class="col-md-8 col-xs-12"> | ||
<h3 class="normal-medium-text">Empowering science!</h3> | <h3 class="normal-medium-text">Empowering science!</h3> | ||
| - | <p>< | + | <p>NEW PLATFORM for distributed computing.</p> |
| - | <p> | + | <p>Volunteers provide the idle calacity of their home computers.</p> |
| - | + | <p>And, with an established user base with more than <span class="larger red-text">1000</span> computers, we effectively bring synthetic biology to society in a new way</p> | |
</div> | </div> | ||
<div class="col-md-4 col-md-offset-0 col-sm-offset-3 col-sm-6 col-xs-offset-2 col-xs-8"> | <div class="col-md-4 col-md-offset-0 col-sm-offset-3 col-sm-6 col-xs-offset-2 col-xs-8"> | ||
Revision as of 14:28, 15 October 2014
 "
"


 CIRCULARIZATION
CIRCULARIZATION
 OLIGOMERIZATION
OLIGOMERIZATION
 FUSION
FUSION
 ON/OFF
ON/OFF
 PURIFICATION
PURIFICATION