Team:ETH Zurich/expresults/rr
From 2014.igem.org
(→Riboregulators) |
(→Riboregulators) |
||
| Line 8: | Line 8: | ||
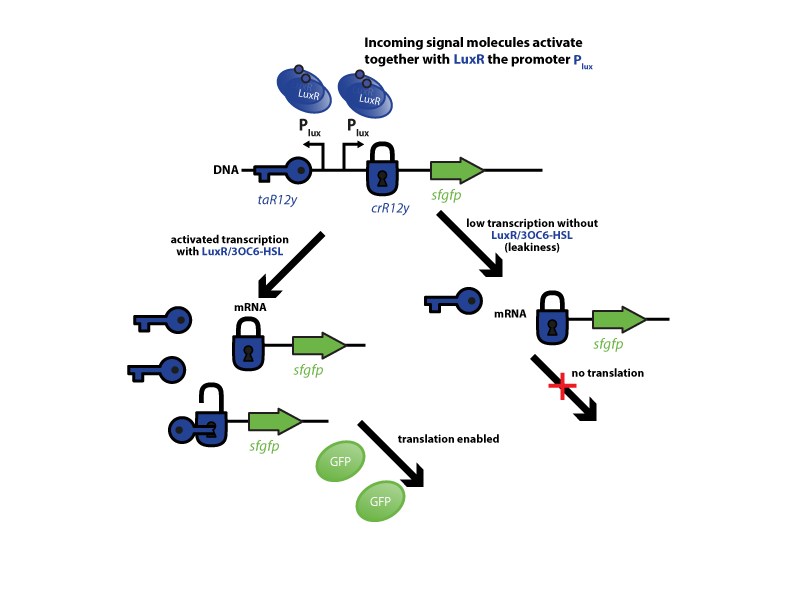
| - | [[File:ETH Zurich2014 Riboregulator expresults.png|center|800px|thumb| '''Figure 1''' Characterization of the riboregulator system. | + | [[File:ETH Zurich2014 Riboregulator expresults.png|center|800px|thumb| '''Figure 1''' Characterization of the riboregulator system. First, a defined amount of the incoming signal molecule 3OC6-HSL binds to LuxR. After dimerization, the complex binds to its corresponding promoter pLux. Both, the ''trans''-activator(taR12y) and the ''cis''-repressor (crR12y) together with superfolder green fluorecent protein (sfGFP), are under control of pLux. Upon induction with LuxR/3OC6-HSL, both are transcribed simultaneously and the taR12y (key symbol) opens up the crR12y sequence (lock) in front of the sfGFP mRNA. This enables translation and sfGFP production (left-hand side). Low transcription (leakiness) gives only small concentrations of mRNA, as a result 'lock' (''cis''-repressor) and 'key' (''trans''-activator) structures do not encounter each other and the translation is not enabled (right-hand side).]] |
The first set of experiments was conducted with cells transformed with two different pPaB plasmids. These plasmids contained either the pBR322 origin (pMB1) or the p15A origin, yielding a stable two-plasmid system with about 15-20 copies per plasmid and cell (medium-copy). Furthermore the published riboregulator sequence includes two forbidden restriction sites (EcoRI and XbaI), since the sites removal could affect the folding or functionality of the part, the original sequences were used as a control. After characterizing the original part the restriction sites were removed by two different approaches: a) multiple site-directed mutagenesis, b) blunting and ligation (Klenow and T4 DNA polymerase). The new Biobrick compatible riboregulator was tested to confirm that no severe loss-of-function occurred due to the site-removal. | The first set of experiments was conducted with cells transformed with two different pPaB plasmids. These plasmids contained either the pBR322 origin (pMB1) or the p15A origin, yielding a stable two-plasmid system with about 15-20 copies per plasmid and cell (medium-copy). Furthermore the published riboregulator sequence includes two forbidden restriction sites (EcoRI and XbaI), since the sites removal could affect the folding or functionality of the part, the original sequences were used as a control. After characterizing the original part the restriction sites were removed by two different approaches: a) multiple site-directed mutagenesis, b) blunting and ligation (Klenow and T4 DNA polymerase). The new Biobrick compatible riboregulator was tested to confirm that no severe loss-of-function occurred due to the site-removal. | ||
In a third step, the construct was transferred into the [http://parts.igem.org/Part:pSB1C3 pSB1C3] backbone (pMB1 origin, high copy number of 100-300) to be in line with the [http://parts.igem.org/Help:Standards/Assembly BioBrick Standard]. Below, you can find the corresponding graphs in figure 2 and figure3, respectively. Future characterization of these constructs should allow us to explore the leakiness dependence on plasmid amount and if the tight expression is robust for different copy numbers. | In a third step, the construct was transferred into the [http://parts.igem.org/Part:pSB1C3 pSB1C3] backbone (pMB1 origin, high copy number of 100-300) to be in line with the [http://parts.igem.org/Help:Standards/Assembly BioBrick Standard]. Below, you can find the corresponding graphs in figure 2 and figure3, respectively. Future characterization of these constructs should allow us to explore the leakiness dependence on plasmid amount and if the tight expression is robust for different copy numbers. | ||
Revision as of 00:08, 18 October 2014
Riboregulators
High basal expression from inactive inducible promoter (leakiness) is a common challenge in the implementation of robust biological control system. The problem is especially severe for systems that approximate digital Boolean logic or drive an amplified downstream response. In the first case an OFF state can be interpreted as ON resulting in incorrect computations. The second scenario leads to high levels of undesired final response. Many former iGEM teams have encountered unwanted basal expression of their genes of interest (goi) due to promoter leakiness (ETH2013, Groningen2012, amongst others), resulting often in narrow optimal operation conditions for their biological devices. Since our system is based on Boolean Logic for its decisions and relies on downstream amplification to regenerate and propagate the signal as a first step we investigated strategies to improve tight gene control. Looking into the available literature we found riboregulators as a promising, highly generalizable approach to address the issue of leakiness[32]. As a convenient proof of concept we coupled the system with quorum sensing modules characterized the response and made the resulting parts available for the iGEM community in the [http://parts.igem.org/Main_Page Registry of Standard Biological Parts]. We suggest that this approach can be generalized to improve many of the inducible system contained in the registry
The riboregulator systems include two parts: 1) a cis-repressed RBS in front of the goi, and 2) a co-expressed trans-activating RNA.
To characterize this system for reliable cell-to-cell communication, we combined the riboregulator part with promoters of the quorum-input-sensing systems used by our team (LuxI/LuxR, LasI/LasR, and RhlI/RhlR). In our final gene circuit, the riboregulators were intended for tight control of integrases. However, to have an easily quantifiable and proportional output, we initially used GFP as our riboregulated goi and measured the output fluorescence with a Tecan plate reader. This approach is described below in the figure 1.
The first set of experiments was conducted with cells transformed with two different pPaB plasmids. These plasmids contained either the pBR322 origin (pMB1) or the p15A origin, yielding a stable two-plasmid system with about 15-20 copies per plasmid and cell (medium-copy). Furthermore the published riboregulator sequence includes two forbidden restriction sites (EcoRI and XbaI), since the sites removal could affect the folding or functionality of the part, the original sequences were used as a control. After characterizing the original part the restriction sites were removed by two different approaches: a) multiple site-directed mutagenesis, b) blunting and ligation (Klenow and T4 DNA polymerase). The new Biobrick compatible riboregulator was tested to confirm that no severe loss-of-function occurred due to the site-removal. In a third step, the construct was transferred into the [http://parts.igem.org/Part:pSB1C3 pSB1C3] backbone (pMB1 origin, high copy number of 100-300) to be in line with the [http://parts.igem.org/Help:Standards/Assembly BioBrick Standard]. Below, you can find the corresponding graphs in figure 2 and figure3, respectively. Future characterization of these constructs should allow us to explore the leakiness dependence on plasmid amount and if the tight expression is robust for different copy numbers.
We have characterized promoters response of the three quorum-sensing systems. These are pLux, pLas, and pRhl. We first measured the transfer functions with a non-regulated RBS in front of sfGFP, i. e. the amount of fluorescence in dependence of the inducer concentration present and compared the response with the corresponding riboregulated constructs with and without the forbidden restriction sites. All experiments were carried out in 96-well microtiter plate format for 10 h. This kinetic data-sets were also used for parameter-fitting in our modeling approach.
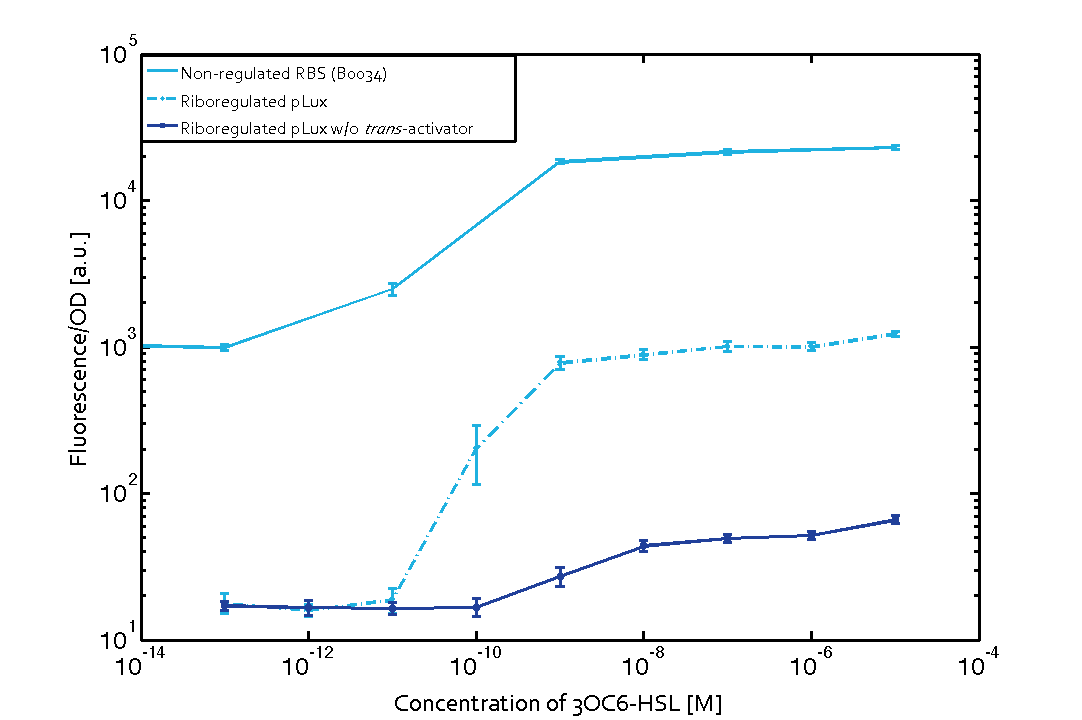
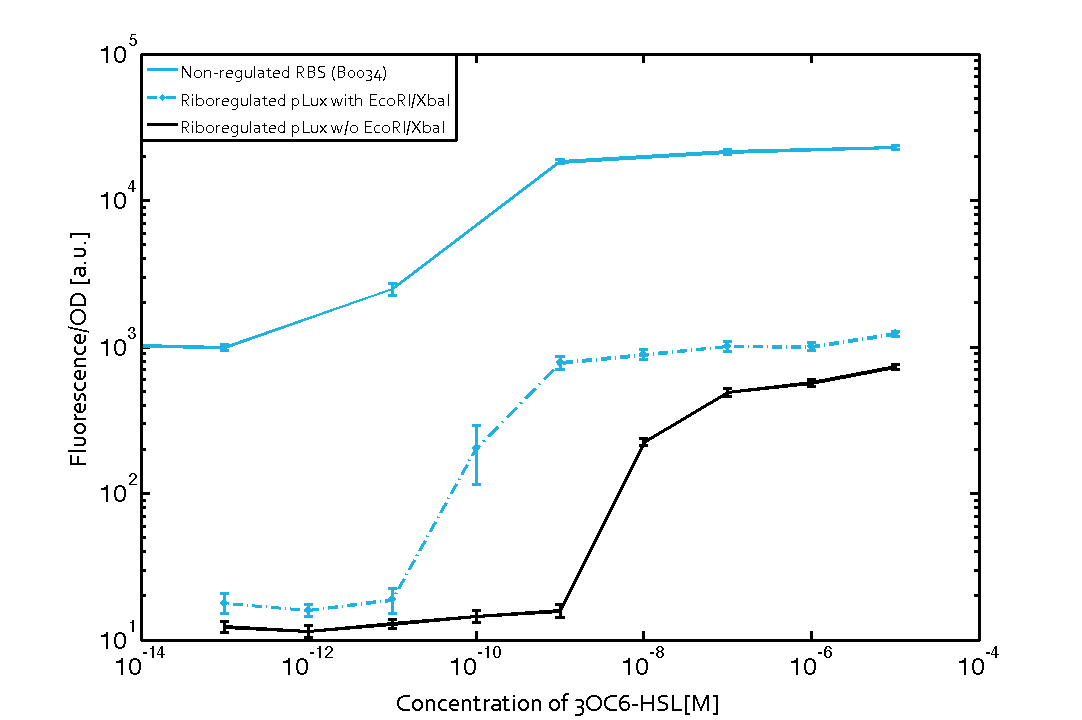
We found an about 60-fold reduced basal GFP expression and an increased signal-to-noise ratio of about 6-fold when using our riboregulator construct, as compared to a non-regulated [http://parts.igem.org/Part:BBa_B0034 reference RBS (B0034)]. These results are summarized in figure 2. The BioBrick conform module was confirmed to show no loss-of-funtion due to EcoRI and XbaI site removal. Interestingly the sensitivity towards the inducer was reduced (see figure 3). Is interesting to speculate if small changes in the sequences flanking the riboregulator could be used as a way to switch the response curve.
| Figure. 2 Improved signal-to-noise ratio and decreased basal GFP expression (leakiness) due to the use of a riboregulator in combination with a quorum-sensing module. The fluorescence per OD600 is shown for the LuxR-system with a complete riboregulator over an inducer-range of 10-13 M to 10-5 M (dashed, light blue). An incomplete riboregulator without the trans-activator shows the expected reduced sensitivity towards the inducer (dark blue). As a reference, a system with a [http://parts.igem.org/Part:BBa_B0034 non-regulated RBS (BBa_B0034)] is shown (light blue). Data points are mean values of triplicate measurements in 96-well microtiter plates 200 min after induction ± standard deviation. For the full data set and kinetics please contact us or visit the raw data page. | Figure. 3 Confirmation of the improved signal-to-noise ratio and decreased basal GFP expression (leakiness) due to the use of a riboregulator without (w/o) EcoRI and XbaI restriction sites in combination with a quorum-sensing module. The fluorescence per OD600 is shown for the LuxR-system with an unchanged riboregulator (dashed, light blue) and a regulator with a changed sequence due to EcoRI and XbaI restriction site removal (dashed, dark blue). The inducer range covers 10-13 M to 10-5 M. As a reference, a system with a [http://parts.igem.org/Part:BBa_B0034 non-regulated RBS (BBa_B0034)] is shown (light blue). Data points are mean values of triplicate measurements in 96-well microtiter plates 200 min after induction ± standard deviation. For the full data set and kinetics please contact us or visit the raw data page. |
 "
"