Team:Austin Texas/kit
From 2014.igem.org
Jordanmonk (Talk | contribs) m |
|||
| (4 intermediate revisions not shown) | |||
| Line 201: | Line 201: | ||
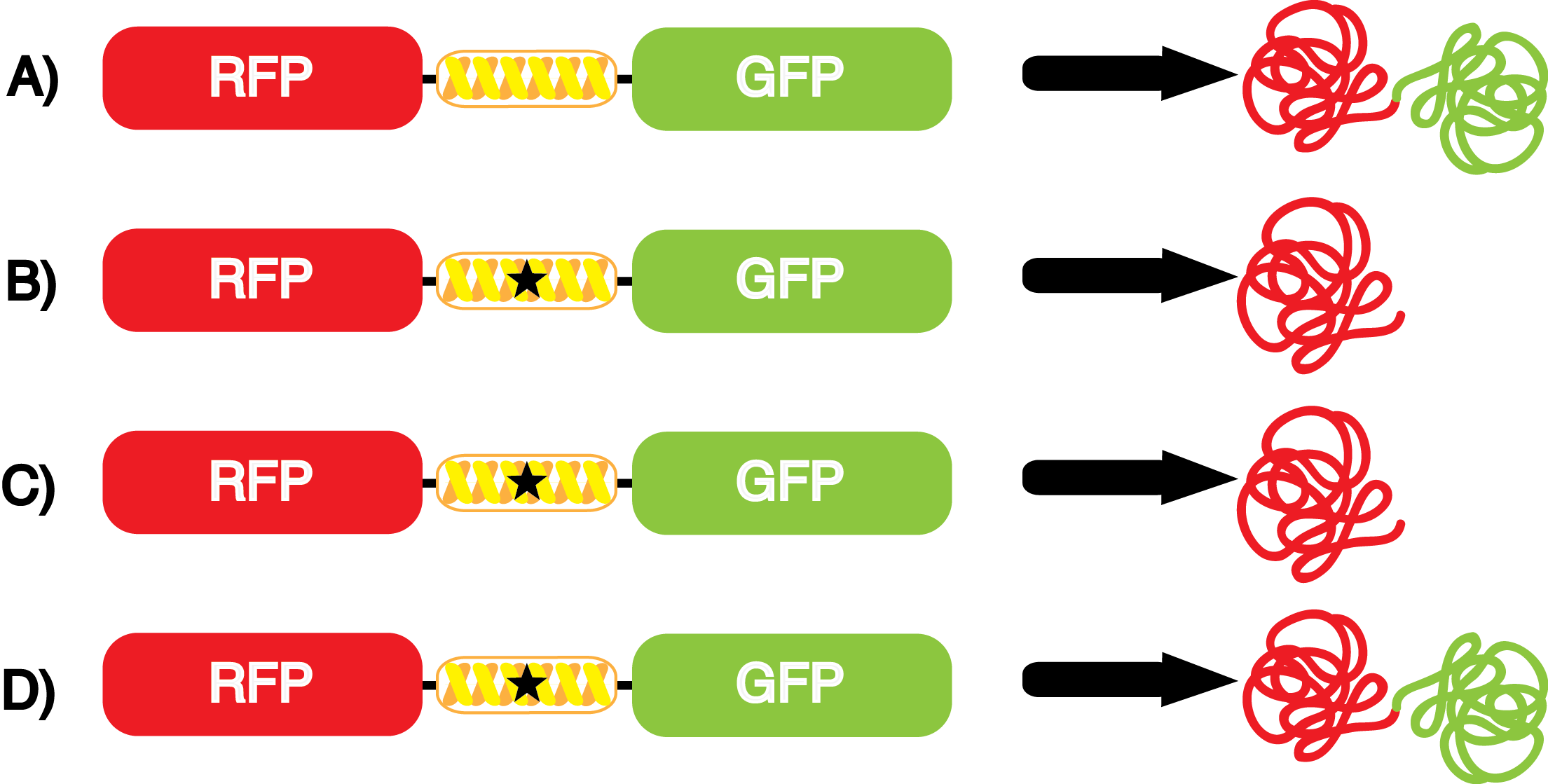
Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they charge their ncAA, ultimately resulting in incorporation of the ncAA in our reporter protein. An inefficient synthetase/tRNA pair will yield incorporation of their ncAA only a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber codon about 10% of the time. | Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they charge their ncAA, ultimately resulting in incorporation of the ncAA in our reporter protein. An inefficient synthetase/tRNA pair will yield incorporation of their ncAA only a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber codon about 10% of the time. | ||
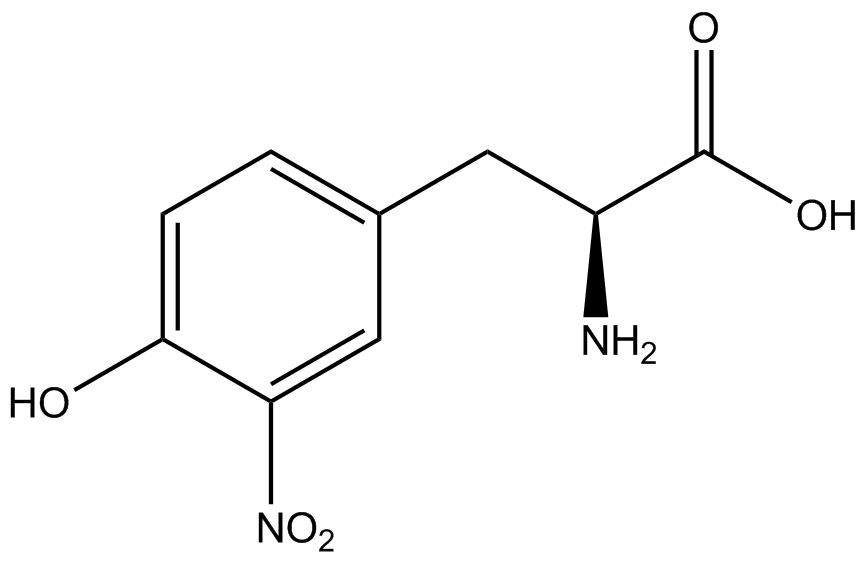
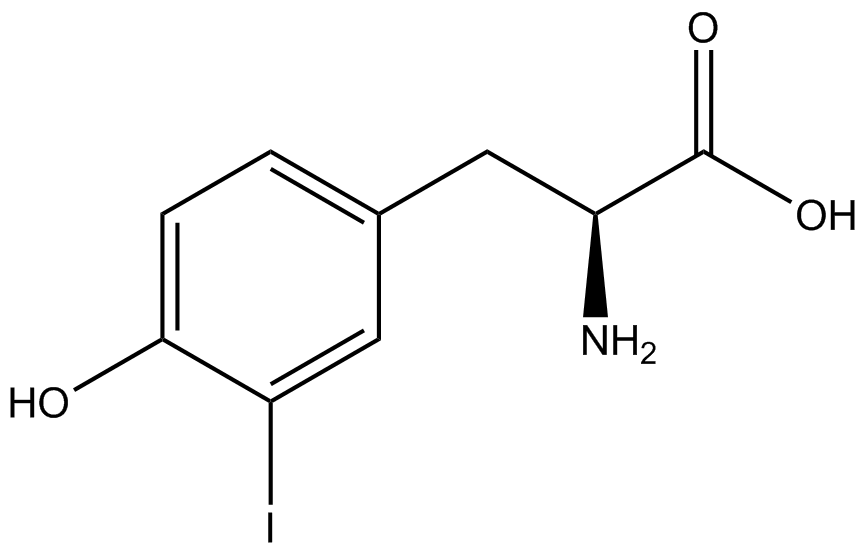
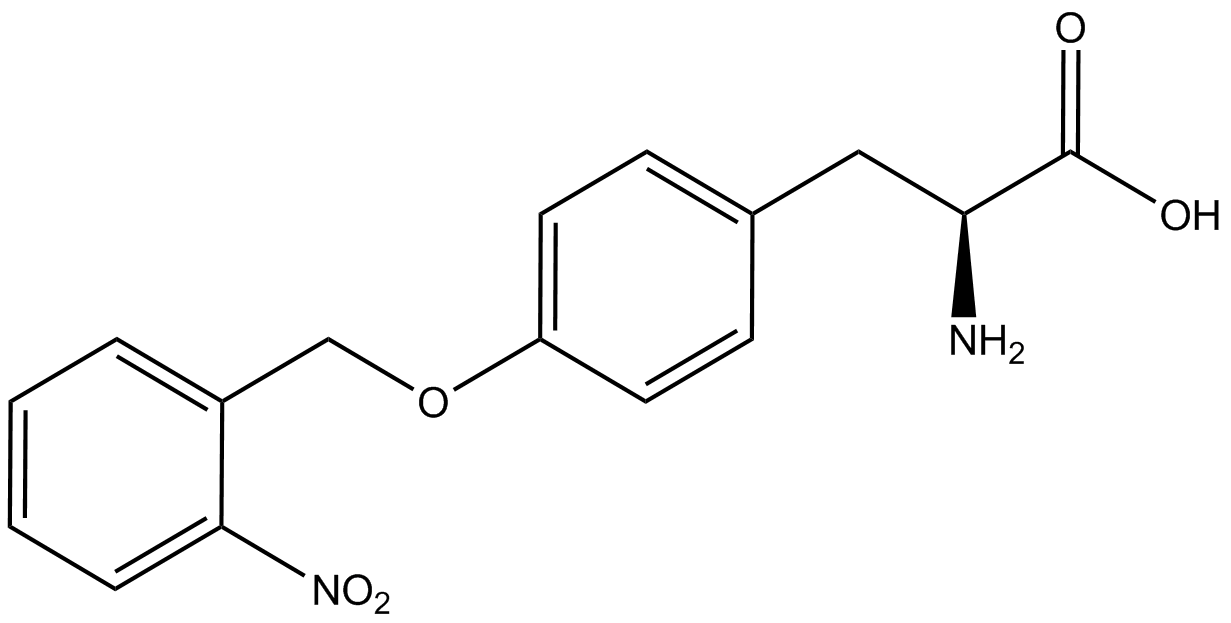
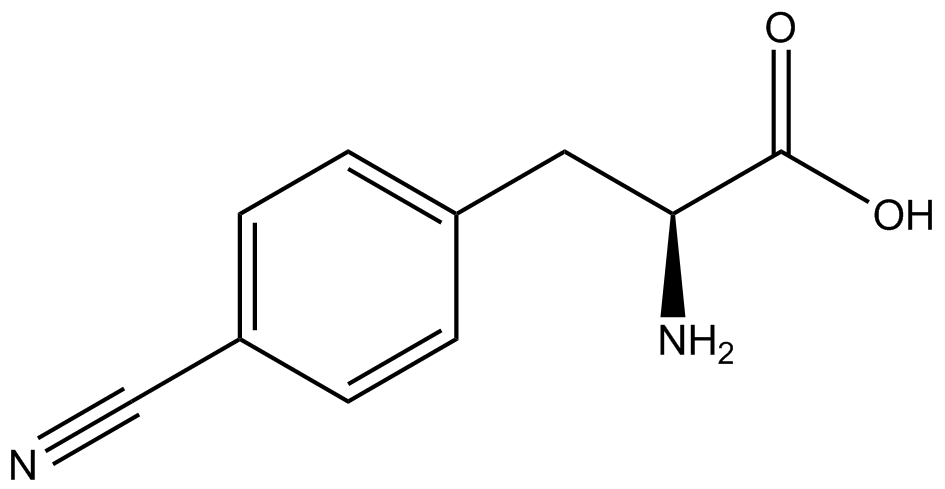
| - | For our results ('''Figure 4'''), two synthetase/tRNA pairs stood out as relatively inefficient: 3- | + | For our results ('''Figure 4'''), two synthetase/tRNA pairs stood out as relatively inefficient: 3-nitro-<small>L</small>-tyrosine and ONBY. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the ncAA was present with the pFRY construct. While both synthetase/tRNA pairs show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitro-<small>L</small>-tyrosine and 20% for ONBY. These results suggest that these synthetase/tRNA pairs do not always efficiently incorporate their ncAA at an amber stop codon. It is important to note that this could partly result from ncAA-related toxicity, and such an effect was repeatedly observed for ONBY. |
<h2>Incorporation Value</h2> | <h2>Incorporation Value</h2> | ||
| - | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|'''Figure 5.''' Incorporation values for each synthetase/tRNA pair. The dashed line equals an incorporation value of 1. Values greater than 1 when using pFRY indicate high efficiency and fidelity of ncAA incorporation. The GFP:RFP relative fluorescence values in the presence and absence of ncAA (see '''Figure 4''') were used to calculate incorporation values. Values were determined by dividing the GFP:RFP relative fluorescence in the presence of ncAA by the GFP:RFP relative fluorescence in the absence of ncAA.]] | + | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|'''Figure 5.''' Incorporation values for each synthetase/tRNA pair. The dashed line equals an incorporation value of 1. Values greater than 1 when using pFRY indicate high efficiency and fidelity of ncAA incorporation. The GFP:RFP relative fluorescence values in the presence and absence of ncAA (see '''Figure 4''') were used to calculate incorporation values. Values were determined by dividing the GFP:RFP relative fluorescence in the presence of ncAA by the GFP:RFP relative fluorescence in the absence of ncAA. Data are presented as the average of three independent cultures. Error bars denote standard deviation.]] |
We wanted a way to better summarize the output of the kit with a single measurement value. We decided to calculate an overall '''incorporation value''' as the GFP:RFP relative fluorescence value in the presence of the ncAA divided by the GFP:RFP relative fluorescence value when the ncAA is not included in the culture. Thus, the incorporation value is a single number that measures the relative efficiency and fidelity of a ncAA tRNA synthetase/tRNA pair. A value of 1 indicates no significant dependence of incorporation on the presence of ncAA in the culture. The higher the value, the higher the fidelity and/or efficiency of the synthetase/tRNA pair. It is important to note that ncAA-related effects on growth and translation can also reduce the apparent incorporation value of a synthetase/tRNA pair below 1. | We wanted a way to better summarize the output of the kit with a single measurement value. We decided to calculate an overall '''incorporation value''' as the GFP:RFP relative fluorescence value in the presence of the ncAA divided by the GFP:RFP relative fluorescence value when the ncAA is not included in the culture. Thus, the incorporation value is a single number that measures the relative efficiency and fidelity of a ncAA tRNA synthetase/tRNA pair. A value of 1 indicates no significant dependence of incorporation on the presence of ncAA in the culture. The higher the value, the higher the fidelity and/or efficiency of the synthetase/tRNA pair. It is important to note that ncAA-related effects on growth and translation can also reduce the apparent incorporation value of a synthetase/tRNA pair below 1. | ||
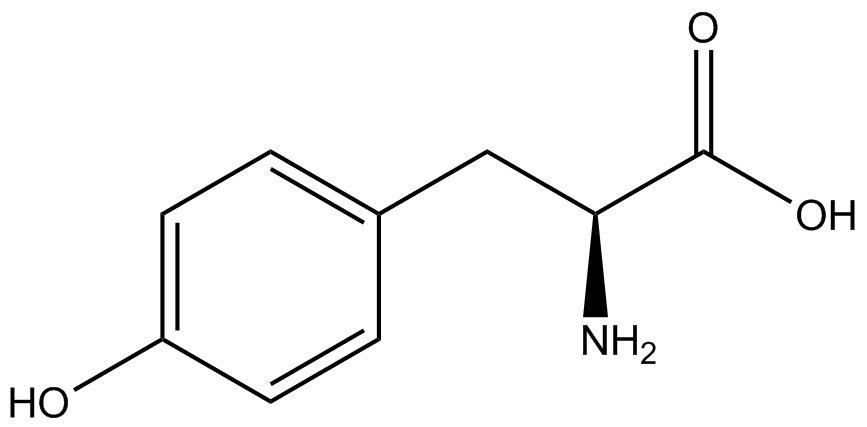
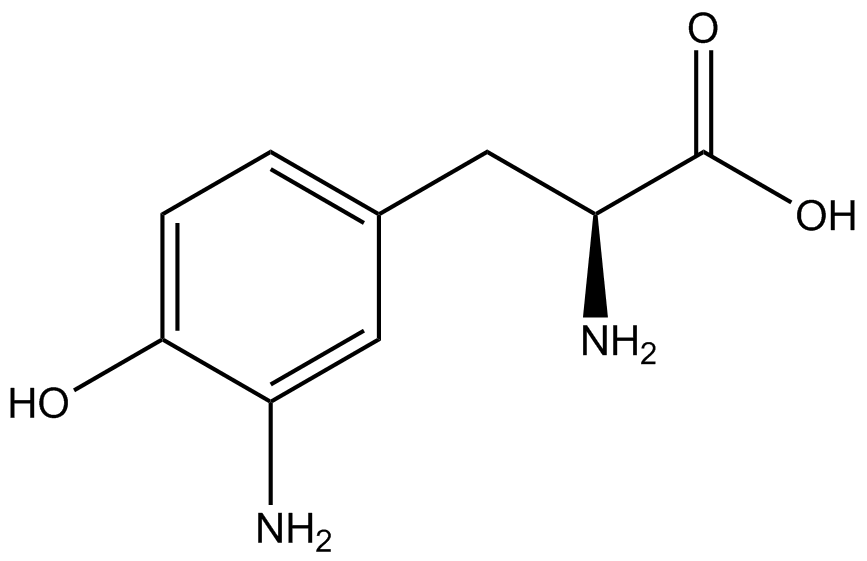
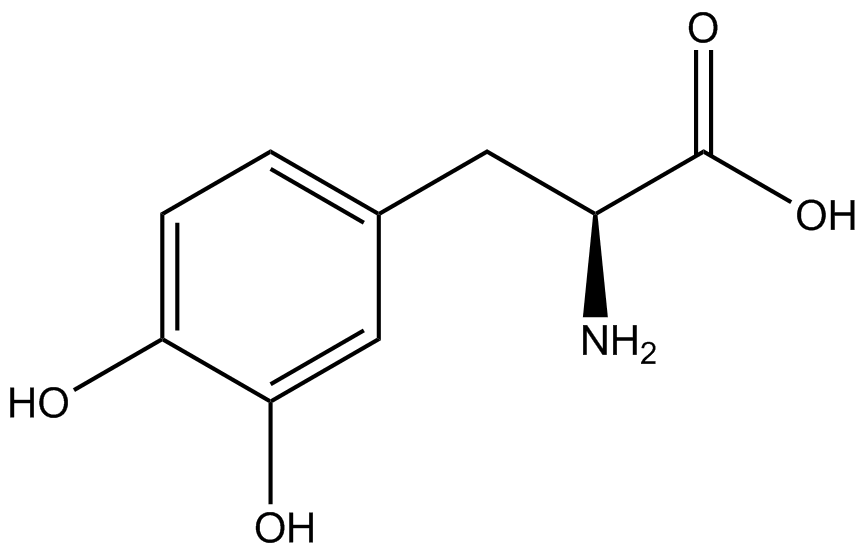
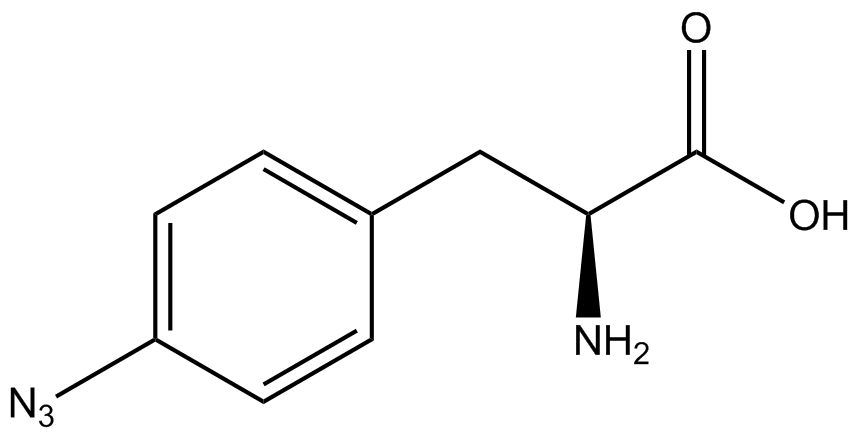
| - | The synthetase/tRNA pairs that showed the highest incorporation values | + | '''The synthetase/tRNA pairs that showed the highest incorporation values were AzF, 3-nitrotyrosine, ONBY, and 3-iodotyrosine.''' Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly (as seen by OD<sub>600</sub> readings) suggesting possible toxicity to the cell. However, ncAA incorporation remained relatively high, resulting in a good incorporation value. '''Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation included 3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and CNF.''' |
<h1>Discussion</h1> | <h1>Discussion</h1> | ||
| - | Our ncAA measurement kit was able to successfully compare the fidelity and efficiency of seven different synthetase/tRNA pairs. We were able to confidently say that one synthetase/tRNA pair had a higher fidelity than another. '''We saw clear evidence that three of our tRNA synthetase/tRNA pairs seemed incapable of discriminating between their ncAA and the 20 canonical amino acids present in media: 3- | + | Our ncAA measurement kit was able to successfully compare the fidelity and efficiency of seven different synthetase/tRNA pairs. We were able to confidently say that one synthetase/tRNA pair had a higher fidelity than another. '''We saw clear evidence that three of our tRNA synthetase/tRNA pairs seemed incapable of discriminating between their ncAA and the 20 canonical amino acids present in media: 3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and CNF.''' We were able to test triplicate cultures of seven different ncAAs and tyrosine (eight total synthetase/tRNA pairs) in one day, generating results that immediately told us which ncAAs worked best in our system both in terms of fidelity and efficiency of incorporation. This required no advanced training or access to equipment besides a shaker and a fluorometer. |
While the measurements may not be perfectly accurate due to the nature of this test, the measurement kit is designed more around ease of use, cost-efficiency, and portability. The ncAA measurement kit can be used to quickly test a synthetase/tRNA pair's quality with minimal effort, as opposed to other more accurate but more intensive or expensive measures such as mass spectrometry. While mass spectrometry can give much more definitive information about a synthetase/tRNA pair, there are many different factors that could make it unreasonable for undergraduate research or high-throughput use. It is a technique that requires a fair amount of skill to do properly, and can be very costly as well. Additionally, the sample needs to be purified from cell culture and prepped for mass spectrometry. | While the measurements may not be perfectly accurate due to the nature of this test, the measurement kit is designed more around ease of use, cost-efficiency, and portability. The ncAA measurement kit can be used to quickly test a synthetase/tRNA pair's quality with minimal effort, as opposed to other more accurate but more intensive or expensive measures such as mass spectrometry. While mass spectrometry can give much more definitive information about a synthetase/tRNA pair, there are many different factors that could make it unreasonable for undergraduate research or high-throughput use. It is a technique that requires a fair amount of skill to do properly, and can be very costly as well. Additionally, the sample needs to be purified from cell culture and prepped for mass spectrometry. | ||
| Line 265: | Line 265: | ||
| 307.09 | | 307.09 | ||
| Soluble in water<br> Heat to 70°C and vortex to dissolve | | Soluble in water<br> Heat to 70°C and vortex to dissolve | ||
| - | | Stock Concentration: 10mM <br>(. | + | | Stock Concentration: 10mM <br>(.03g in 10mL H2O)<br> Concentration in Culture: 1mM |
|- | |- | ||
Latest revision as of 23:54, 18 March 2015
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"