Team:Austin Texas/kit
From 2014.igem.org
| Line 130: | Line 130: | ||
The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient at conducting every step in the process of reading through an amber codon.]] | The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient at conducting every step in the process of reading through an amber codon.]] | ||
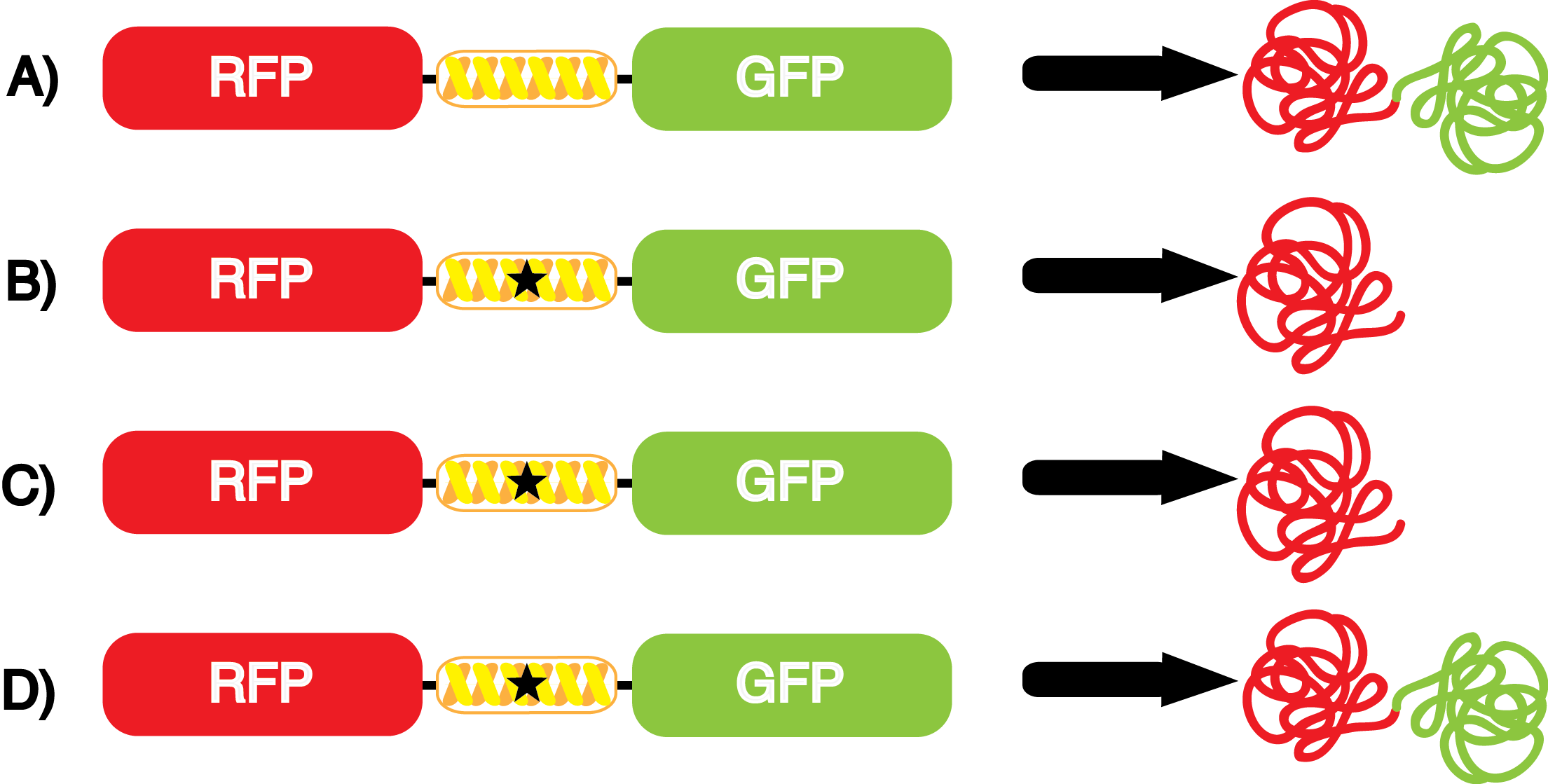
| - | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence (Figure 3A). This happens because the reporter contains no amber codon, and thus does not require a ncAA synthetase/tRNA pair. | + | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence ('''Figure 3A'''). This happens because the reporter contains no amber codon, and thus does not require a ncAA synthetase/tRNA pair. |
| - | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence (Figure 3B). This occurs because without the pStG, the amber codon is not recoded to allow for ncAA incorporation. | + | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence ('''Figure 3B'''). This occurs because without the pStG, the amber codon is not recoded to allow for ncAA incorporation. |
| - | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence (Figure 3C). This occurs because while the codon has been recoded, there is no ncAA and without ncAA there can be no incorporation at the amber codon. | + | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence ('''Figure 3C'''). This occurs because while the codon has been recoded, there is no ncAA and without ncAA there can be no incorporation at the amber codon. |
| - | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence (Figure 3D). This occurs because the synthetase found in pStG can covalently attach the ncAA to the tRNA gene present in pStG. This "charged tRNA" is then present during translation, allowing incorporation of the ncAA into the reporter protein. | + | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence ('''Figure 3D'''). This occurs because the synthetase found in pStG can covalently attach the ncAA to the tRNA gene present in pStG. This "charged tRNA" is then present during translation, allowing incorporation of the ncAA into the reporter protein. |
| Line 191: | Line 191: | ||
We wanted to test whether the ncAA synthetase/tRNA pairs would incorporate anything besides their amino acid at the amber stop codon (UAG), and that they would in fact incorporate their specific ncAA if it was present. This is essentially the fidelity of each ncAA sythetase/tRNA pair. | We wanted to test whether the ncAA synthetase/tRNA pairs would incorporate anything besides their amino acid at the amber stop codon (UAG), and that they would in fact incorporate their specific ncAA if it was present. This is essentially the fidelity of each ncAA sythetase/tRNA pair. | ||
| - | Fidelity was measured by comparing the production of GFP in cultures containing pStG/pFRY with or without ncAA. In the absence of a ncAA only RFP should be translated, as translation is expected to halt between RFP and GFP at the amber codon on the linker sequence in pFRY. Alternatively, if the corresponding ncAA for pStG is present or if the synthetase/tRNA pair has low fidelity and can misincorporate a different amino acid, translation should continue through the UAG. In this case, RFP and GFP should both be translated. We also tested pStG/pFRYC strains in (+/-) ncAA conditions as a control for what effect the ncAA has on the fluorescence or growth of the cells. These strains should express RFP and GFP in all conditions since pFRYC does not have an amber codon in the linker (Figure 2). However, if the presence of ncAA affects cell growth or fluorescence activity, we will need these controls to determine the extent of the effect. | + | Fidelity was measured by comparing the production of GFP in cultures containing pStG/pFRY with or without ncAA. In the absence of a ncAA only RFP should be translated, as translation is expected to halt between RFP and GFP at the amber codon on the linker sequence in pFRY. Alternatively, if the corresponding ncAA for pStG is present or if the synthetase/tRNA pair has low fidelity and can misincorporate a different amino acid, translation should continue through the UAG. In this case, RFP and GFP should both be translated. We also tested pStG/pFRYC strains in (+/-) ncAA conditions as a control for what effect the ncAA has on the fluorescence or growth of the cells. These strains should express RFP and GFP in all conditions since pFRYC does not have an amber codon in the linker ('''Figure 2'''). However, if the presence of ncAA affects cell growth or fluorescence activity, we will need these controls to determine the extent of the effect. |
To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD<sub>600</sub> of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. | To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD<sub>600</sub> of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. | ||
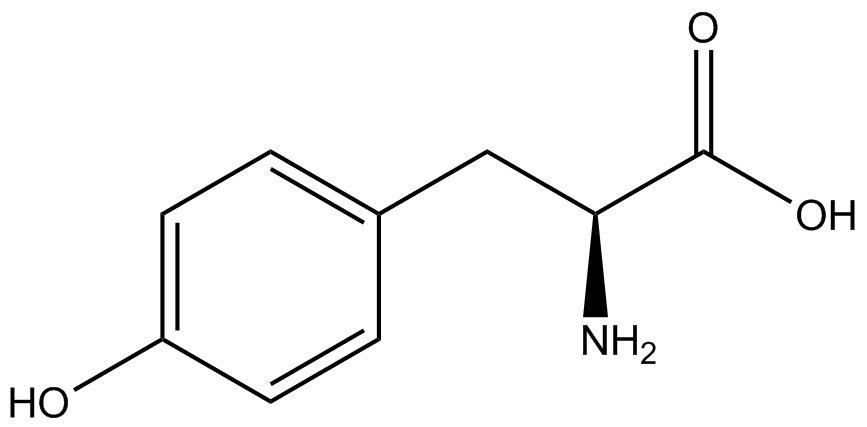
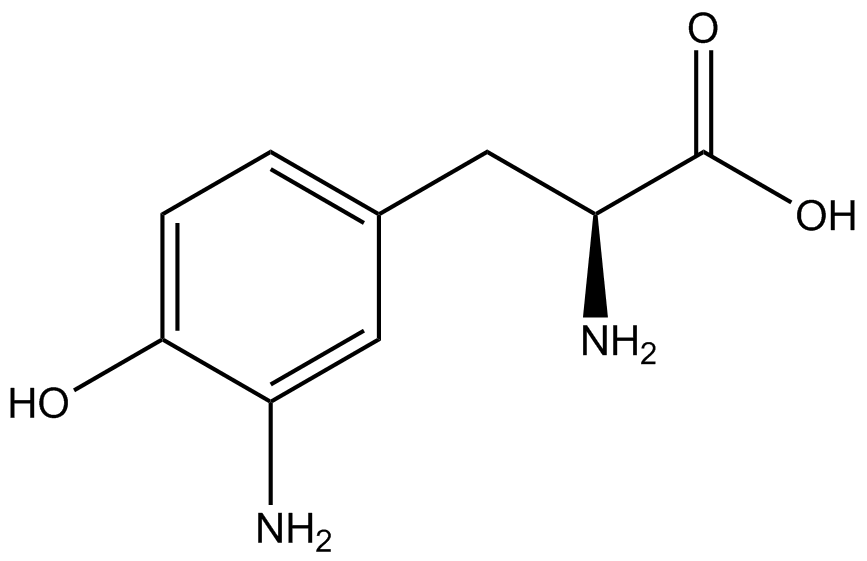
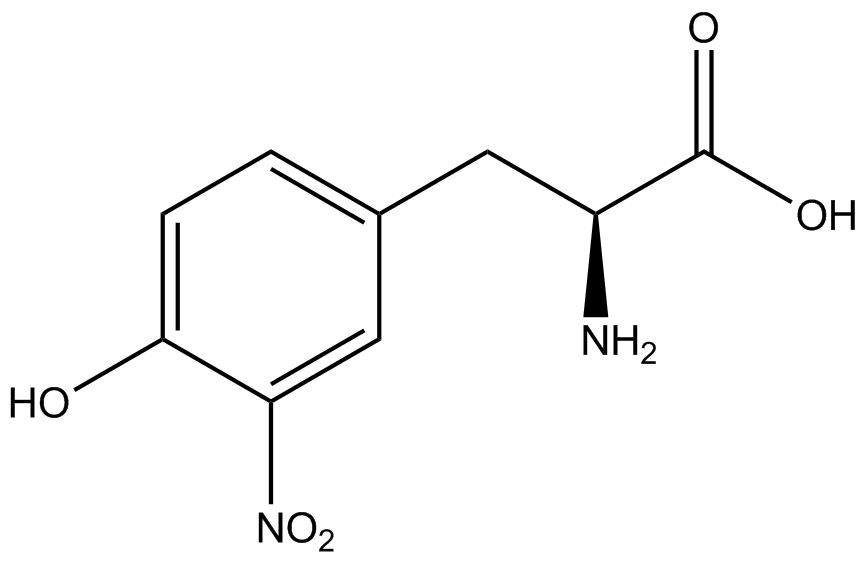
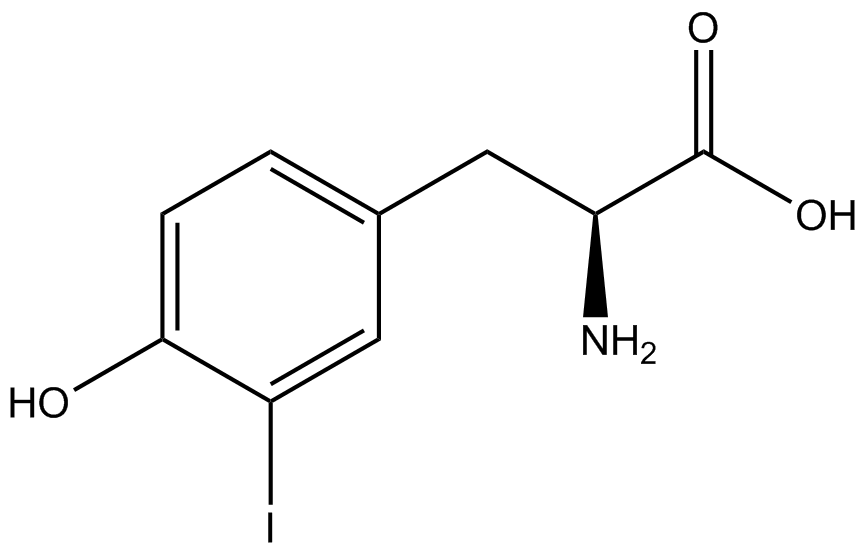
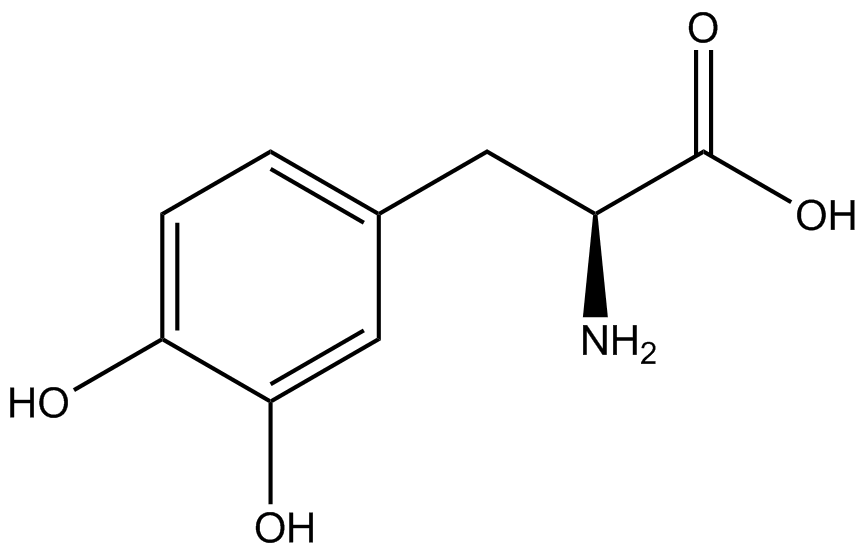
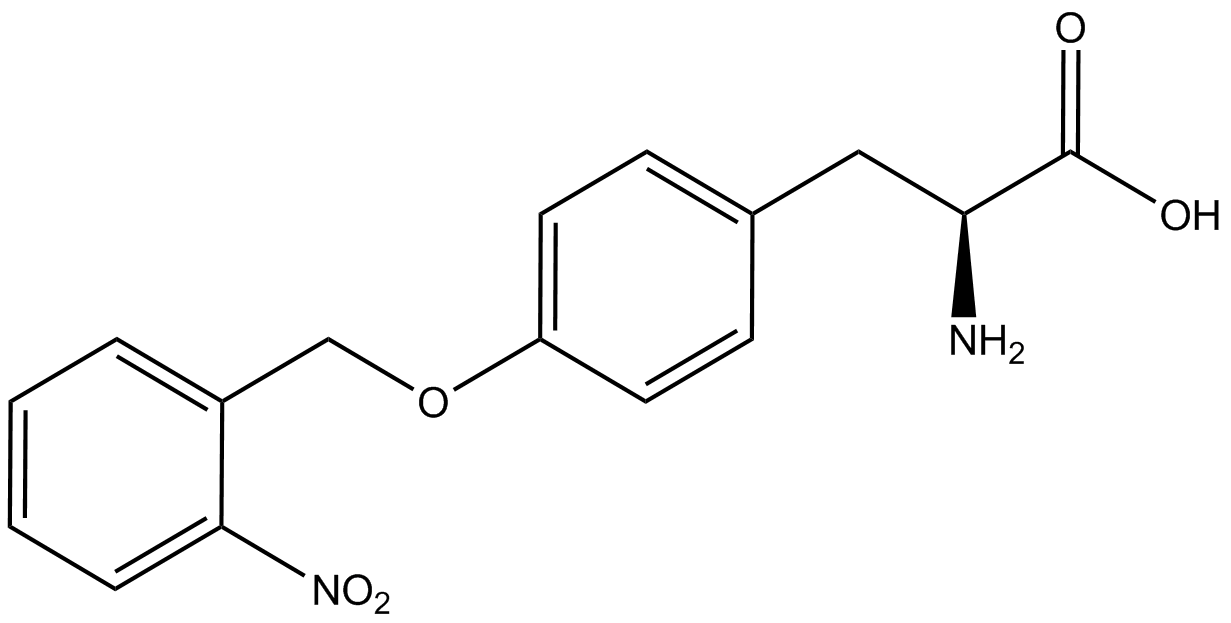
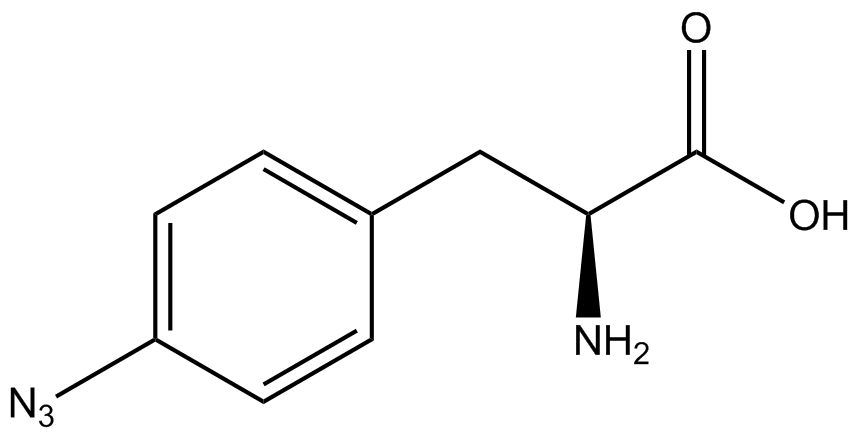
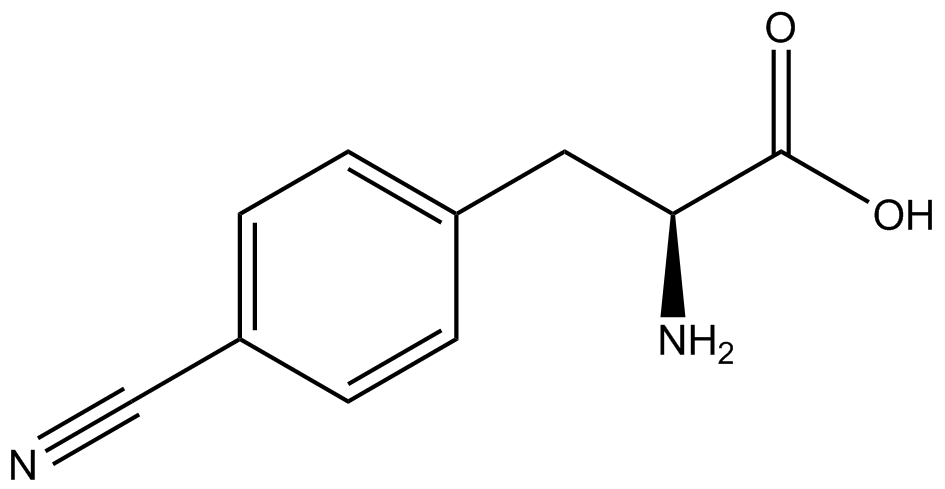
| - | When these values were graphed (Figure 4), some synthetase/tRNA pairs such as 4-azido-<small>L</small>-phenylalanine (AzF), 3-nitro-<small>L</small>-tyrosine, 3-iodo-<small>L</small>-tyrosine, and ''o''-(2-nitrobenzyl)-<small>L</small>-tyrosine (ONBY) resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and 4-cyano-<small>L</small>-phenylalanine (CNF)) did not show a difference in GFP fluorescence normalized to RFP fluorescence dependent on the presence of ncAA, indicating that these pairs incorporated other amino acids at the amber codon when their specific ncAA was not present (and perhaps even when it was), and thus have a low fidelity. | + | When these values were graphed ('''Figure 4'''), some synthetase/tRNA pairs such as 4-azido-<small>L</small>-phenylalanine (AzF), 3-nitro-<small>L</small>-tyrosine, 3-iodo-<small>L</small>-tyrosine, and ''o''-(2-nitrobenzyl)-<small>L</small>-tyrosine (ONBY) resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-amino-<small>L</small>-tyrosine, <small>L</small>-DOPA, and 4-cyano-<small>L</small>-phenylalanine (CNF)) did not show a difference in GFP fluorescence normalized to RFP fluorescence dependent on the presence of ncAA, indicating that these pairs incorporated other amino acids at the amber codon when their specific ncAA was not present (and perhaps even when it was), and thus have a low fidelity. |
<h2>Synthetase Efficiency</h2> | <h2>Synthetase Efficiency</h2> | ||
| Line 201: | Line 201: | ||
Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they charge their ncAA, ultimately resulting in incorporation of the ncAA in our reporter protein. An inefficient synthetase/tRNA pair will yield incorporation of their ncAA only a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber codon about 10% of the time. | Another measure of quality for these ncAA synthetase/tRNA pairs is how efficiently they charge their ncAA, ultimately resulting in incorporation of the ncAA in our reporter protein. An inefficient synthetase/tRNA pair will yield incorporation of their ncAA only a fraction of the time, even when their ncAA is present. Our system can be used to measure this level of efficiency, though it does not indicate why a particular synthetase is efficient or inefficient. By comparing the level of GFP fluorescence to the normalized level of RFP fluorescence when the amino acid is present, we can see how efficient the synthetase is. In essence, if the normalized fluorescence of GFP relative to RFP is close to 100% (if the GFP is expressed roughly 100% of the time that RFP is expressed), then the synthetase is very efficient. On the other side, if say the normalized fluorescence of GFP relative to RFP is closer to 10%, then the synthetase would not be very efficient, because even when the ncAA was there, it only incorporated it at the amber codon about 10% of the time. | ||
| - | For our results (Figure 4), two synthetase/tRNA pairs stood out as relatively inefficient: 3-nitrotyrosine and ONBY. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the ncAA was present with the pFRY construct. While both synthetase/tRNA pairs show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitrotyrosine and 20% for ONBY. These results suggest that | + | For our results ('''Figure 4'''), two synthetase/tRNA pairs stood out as relatively inefficient: 3-nitrotyrosine and ONBY. Both of these synthetase/tRNA pairs showed a significantly smaller normalized GFP to RFP fluorescence when the ncAA was present with the pFRY construct. While both synthetase/tRNA pairs show a significant increase in normalized GFP to RFP fluorescence when the amino acid was present compared to when it was absent, which indicates a high fidelity, the actual GFP fluorescence relative to the RFP fluorescence was only around 50% for 3-nitrotyrosine and 20% for ONBY. These results suggest that these synthetase/tRNA pairs do not always efficiently incorporate their ncAA at an amber stop codon. It is important to note that this could partly result from ncAA-related toxicity, and such an effect was repeatedly observed for ONBY. |
<h2>Incorporation Value</h2> | <h2>Incorporation Value</h2> | ||
| - | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|'''Figure 5.''' Incorporation values for each synthetase/tRNA pair. | + | [[File:UT_Austin_2014_Kit_Incorporation_Value_Graph.png|600px|thumb|'''Figure 5.''' Incorporation values for each synthetase/tRNA pair. The dashed line equals an incorporation value of 1. Values greater than 1 when using pFRY are desirable. The GFP:RFP relative fluorescence values in the presence and absence of ncAA (see '''Figure 4''') were used to calculate incorporation values. Values were determined by dividing the GFP:RFP relative fluorescence in the presence of ncAA by the GFP:RFP relative fluorescence in the absence of ncAA. This value does not correct for ncAA-related effects on translation, which can occasionally be seen with the pFRYC controls.]] |
| - | The incorporation value is a single number that measure the relative efficiency and fidelity of a ncAA tRNA synthetase/tRNA pair. A value of 1 or lower indicates no significant incorporation dependent on the presence of ncAA in the culture. The higher the value, the higher the fidelity and/or efficiency of the synthetase/tRNA pair. It is important to note that ncAA-related effects on growth and translation can reduce the incorporation value of a synthetase/tRNA pair | + | The incorporation value is a single number that measure the relative efficiency and fidelity of a ncAA tRNA synthetase/tRNA pair. A value of 1 or lower indicates no significant incorporation dependent on the presence of ncAA in the culture. The higher the value, the higher the fidelity and/or efficiency of the synthetase/tRNA pair. It is important to note that ncAA-related effects on growth and translation can reduce the incorporation value of a synthetase/tRNA pair. |
The synthetase/tRNA pairs that showed the highest incorporation values include AzF, 3-nitrotyrosine, ONBY, and 3-iodotyrosine in decreasing order. Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly (as seen by OD600 readings, data not shown) suggesting possible toxicity to the cell. However, ncAA incorporation remained relatively high, resulting in a good incorporation value. Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation include 3-aminotyrosine, <small>L</small>-DOPA, and CNF. | The synthetase/tRNA pairs that showed the highest incorporation values include AzF, 3-nitrotyrosine, ONBY, and 3-iodotyrosine in decreasing order. Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly (as seen by OD600 readings, data not shown) suggesting possible toxicity to the cell. However, ncAA incorporation remained relatively high, resulting in a good incorporation value. Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation include 3-aminotyrosine, <small>L</small>-DOPA, and CNF. | ||
Revision as of 02:27, 18 October 2014
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"