Team:Austin Texas/kit
From 2014.igem.org
| Line 102: | Line 102: | ||
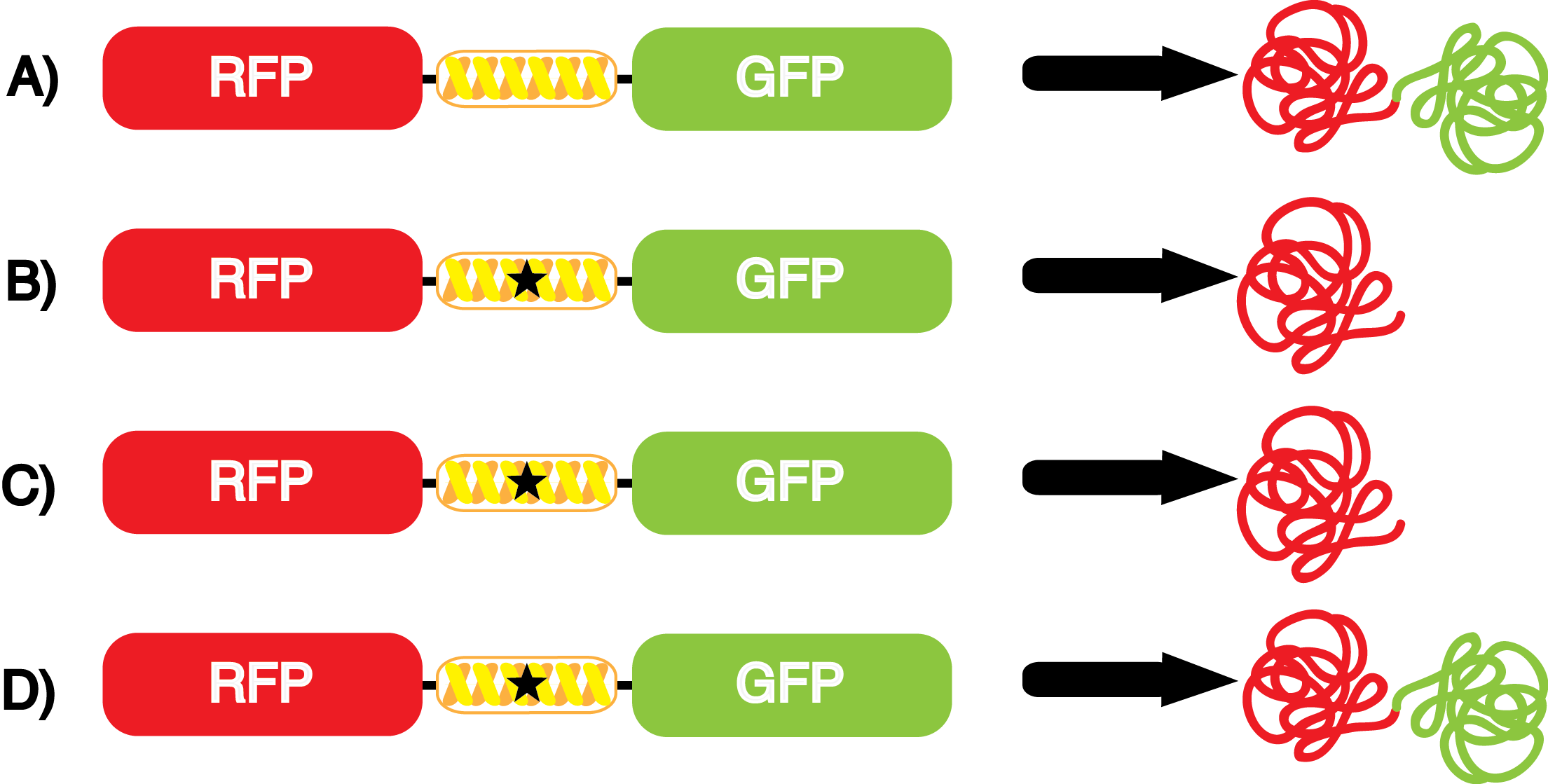
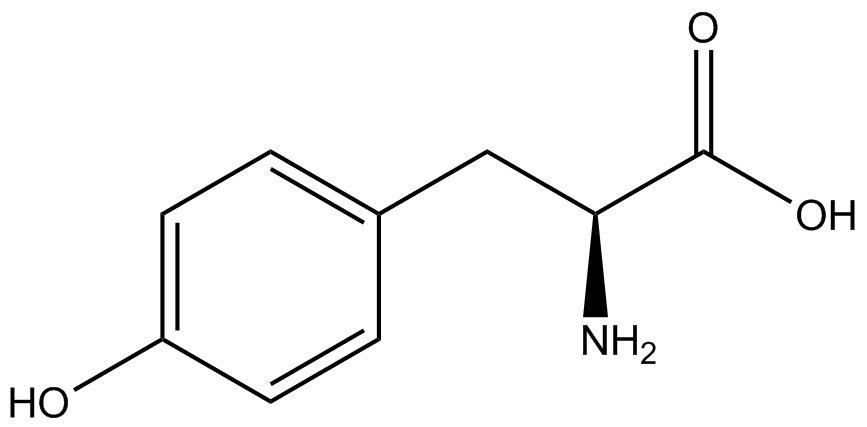
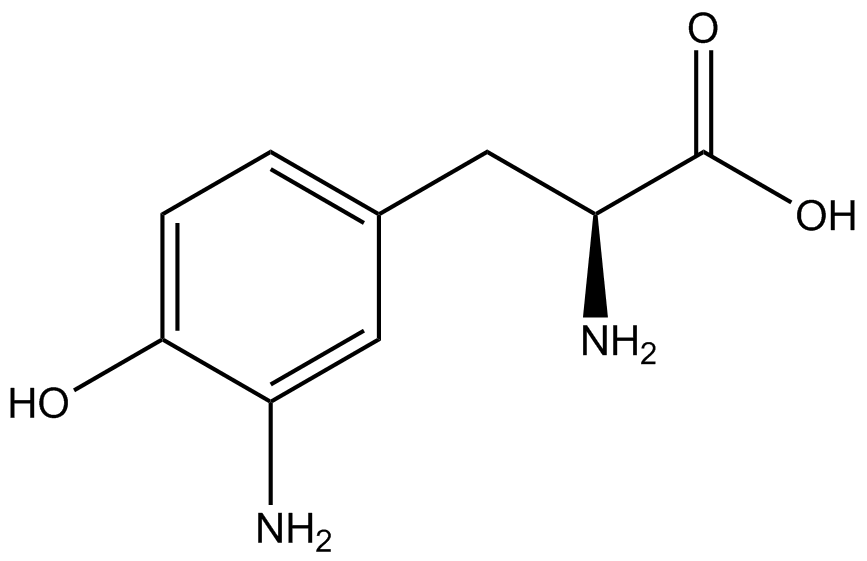
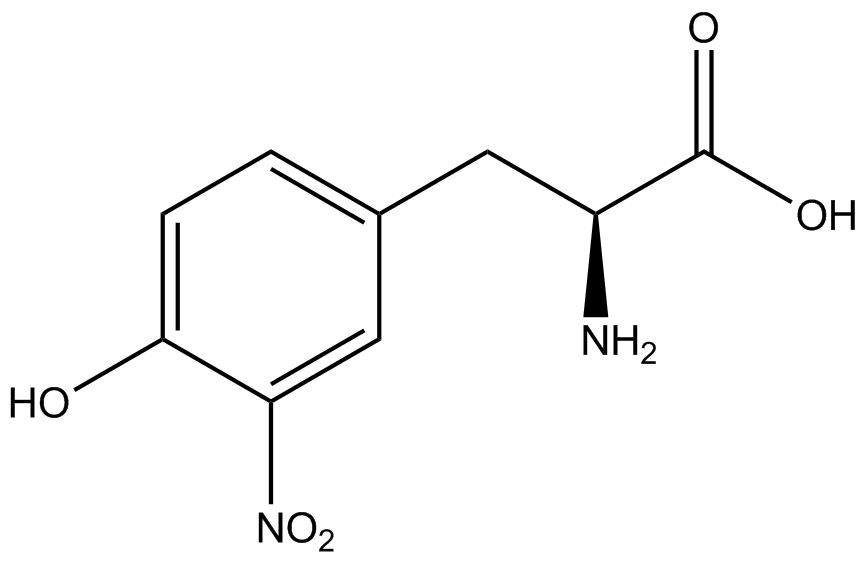
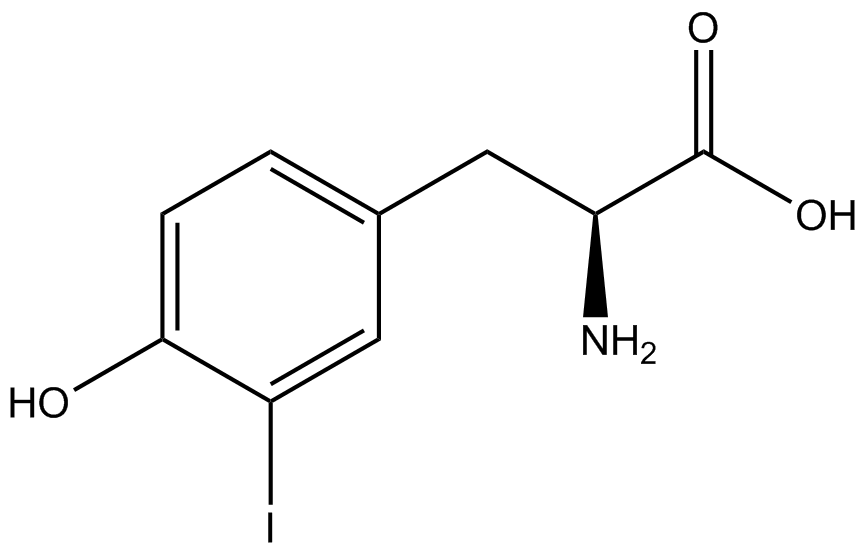
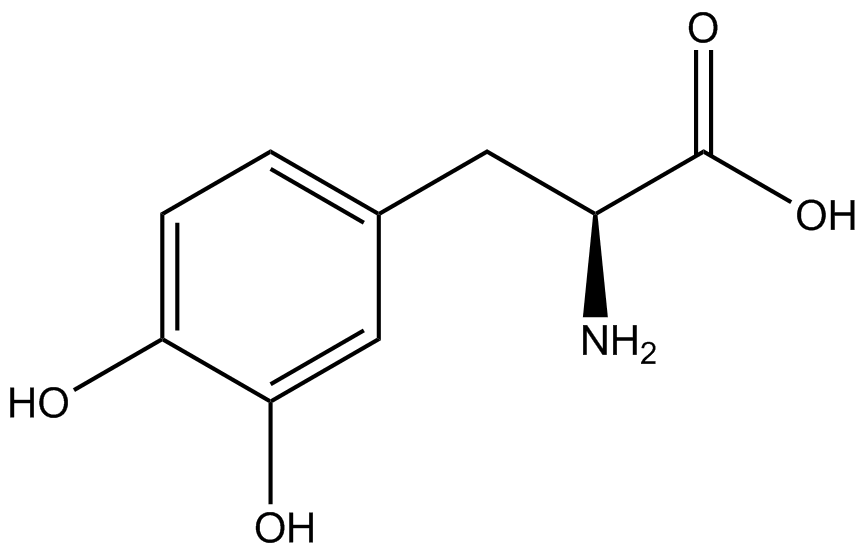
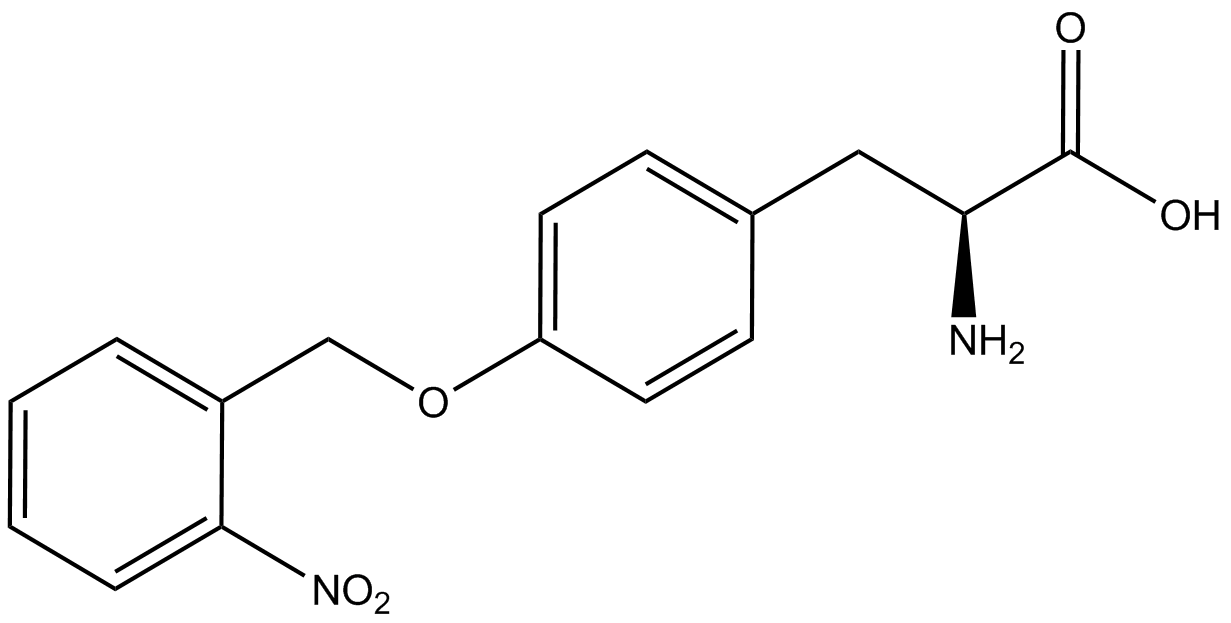
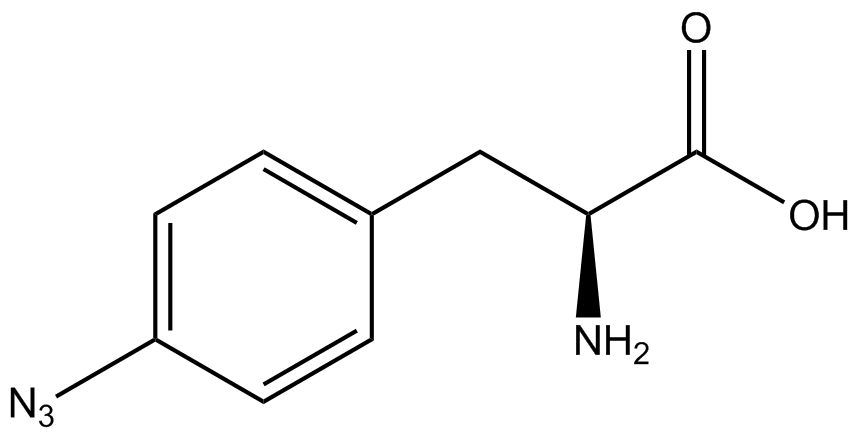
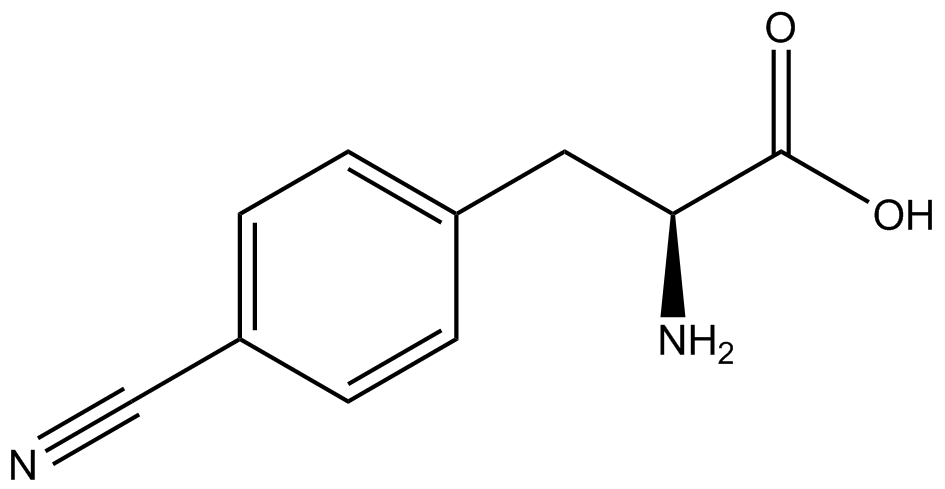
| - | The Shultz lab was the first to expand the genetic code using a unique synthetase/tRNA. This synthetase /tRNA pair originated from M. jannaschii tyrosine RS (Mj-TyrRS) and allowed the cell to incorporate <i>O</i>-methyl-L-tyrosine at the amber codon. Since then, several ncAA synthetase/pairs have been generated. A total of seven ncAAs were used in addition to tyrosine for our project. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. | + | The Shultz lab was the first to expand the genetic code using a unique synthetase/tRNA. This synthetase /tRNA pair originated from M. jannaschii tyrosine RS (Mj-TyrRS) and allowed the cell to incorporate <i>O</i>-methyl-L-tyrosine at the amber codon (Wang et al. 2001). Since then, several ncAA synthetase/tRNA pairs have been generated. A total of seven ncAAs were used in addition to tyrosine for our project. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. |
| - | + | It is important to note that complications can arise when the genetic code is recoded. In a normal bacterium, release factor RF1 is responsible for terminating translation when the ribosome reaches an amber stop codon. To avoid termination at a UAG amber codon, a strain of ''E. coli'' was engineered by the Church and Isaacs groups using MAGE and CAGE to remove all of the amber codons from the genome and knock out the RF1 gene (Isaacs et al. 2011). The resulting strain, called "amberless" ''E. coli'', has NO AMBER CODONS. Thus, to better characterize the synthetase/tRNA pairs, we have employed amberless ''E. coli''. The advantage of this system is that since there are no amber codons in the genome, we cannot accidentally interfere with any cellular processes. Additionally, since RF1 is knocked out, there is no competition between RF1 and the novel synthetase/tRNA pair. These factors allow us to better assess the fidelity and efficiency of ncAA tRNA synthetase/tRNA pairs. | |
<h1>Experimental Design and Method</h1> | <h1>Experimental Design and Method</h1> | ||
Revision as of 16:43, 17 October 2014
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"