Team:Austin Texas/kit
From 2014.igem.org
| Line 222: | Line 222: | ||
<h1>Discussion</h1> | <h1>Discussion</h1> | ||
| - | |||
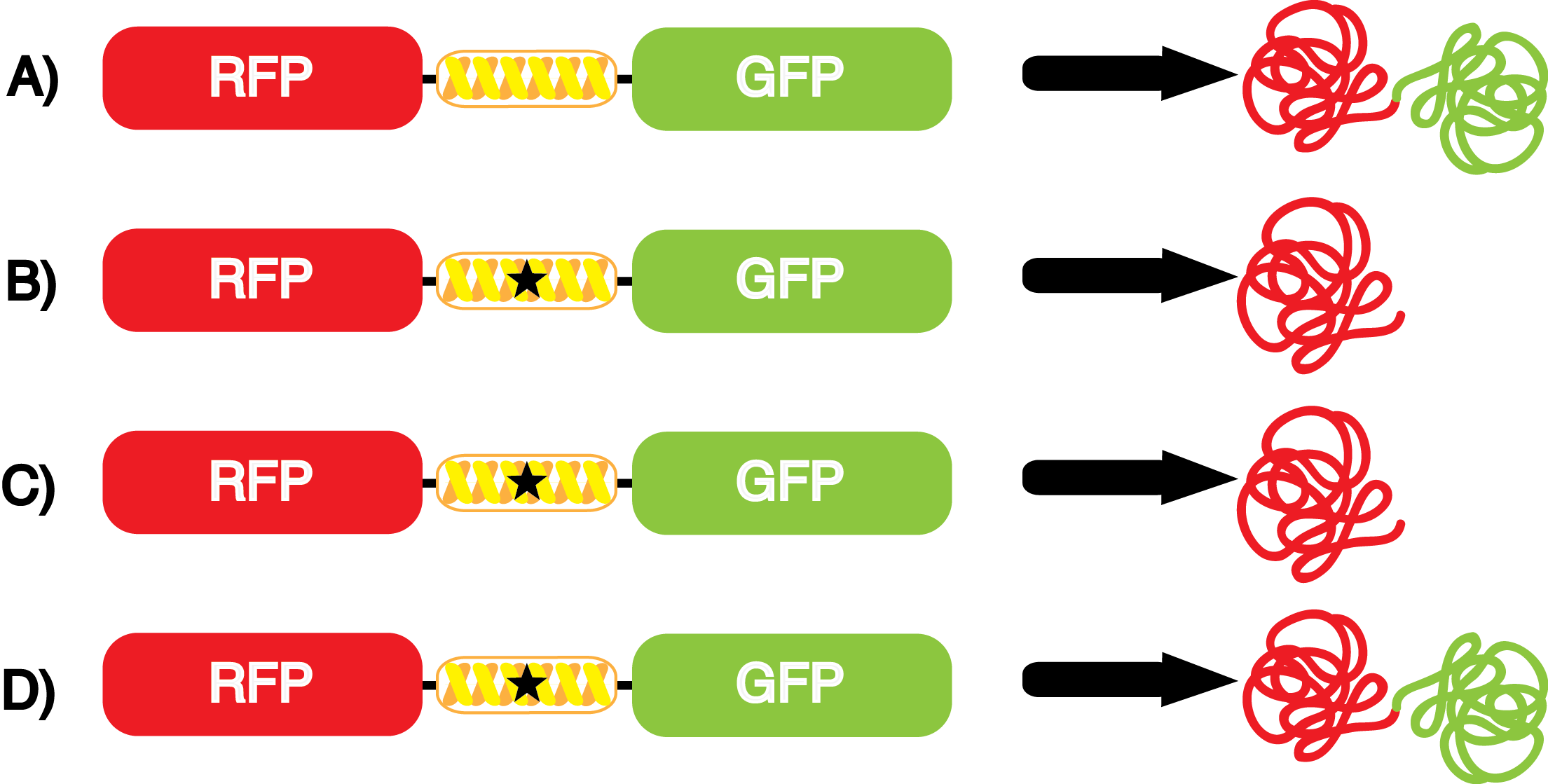
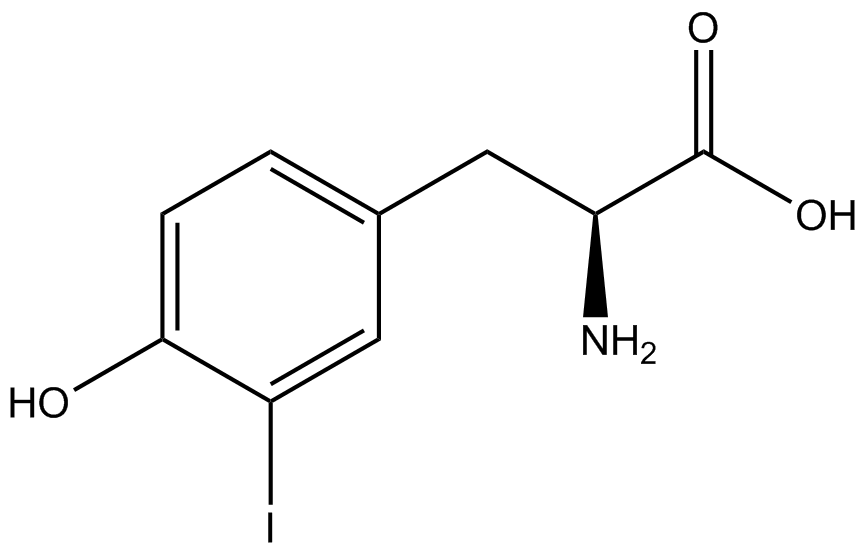
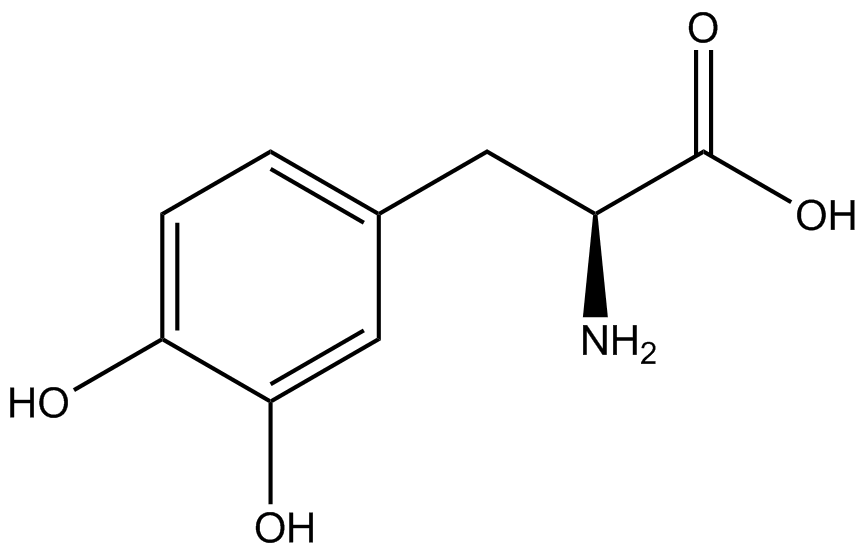
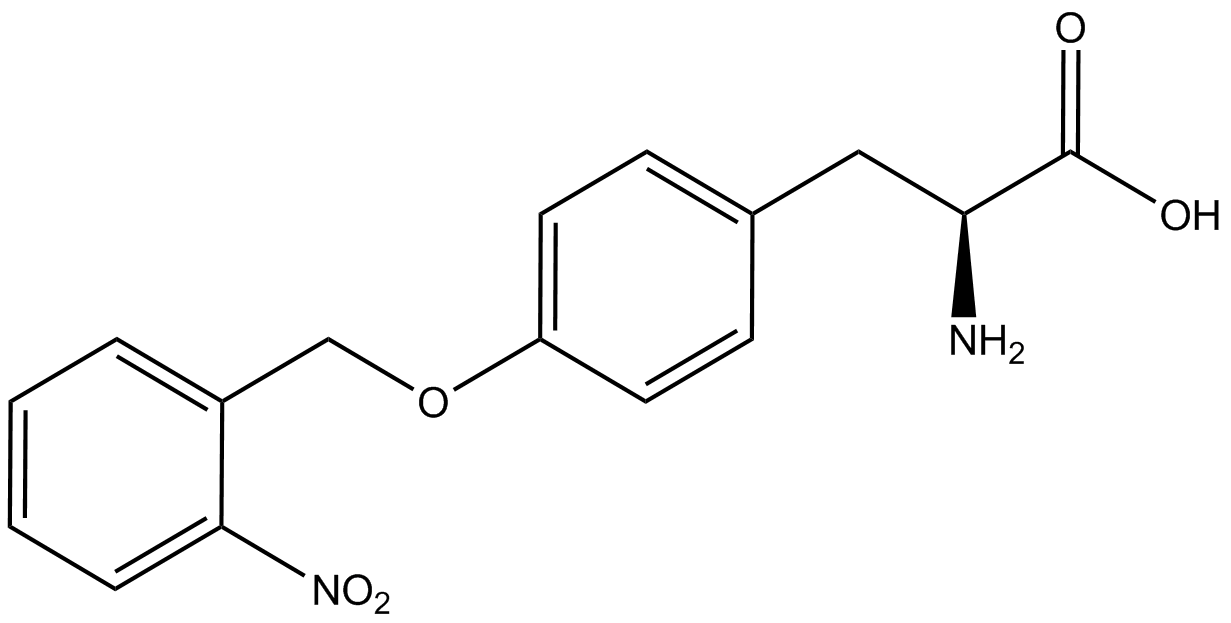
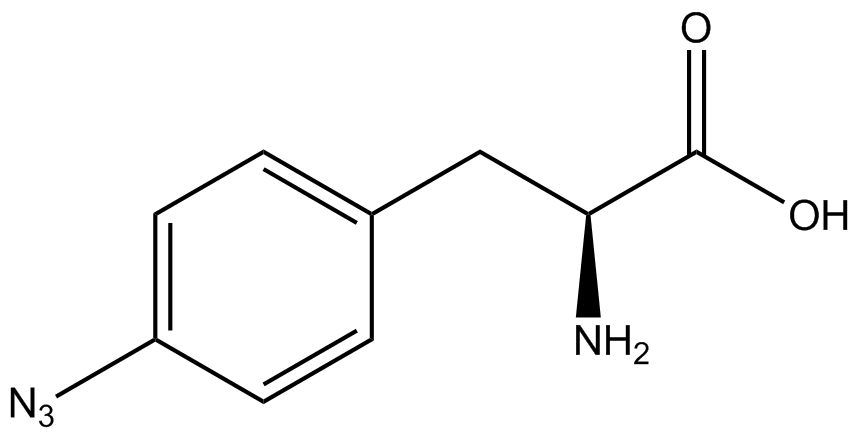
Our ncAA measurement kit was able to successfully compare the fidelity and efficiency of seven different synthetase/tRNA pairs. We were able to confidently say that one synthetase/tRNA pair had a higher fidelity than another. While the measurements may not be perfectly accurate due to the nature of this test, the measurement kit is designed more around ease of use, cost-efficiency, and portability. The ncAA measurement kit can be used to quickly test a synthetase/tRNA pair's quality with minimal effort, as opposed to other more accurate but more intensive or expensive measures such as mass spectrometry. While mass spectrometry can give much more definitive information about a synthetase/tRNA pair, there are many different factors that could make it unreasonable for undergraduate research. It is a technique that requires a fair amount of skill to do properly, and can be very costly as well. For the ncAA measurement kit, all that is necessary is to order the kit parts (pFRYC and PFRY in separate strains of amberless ''E. coli'') from the iGEM registry, transform the synthetase/tRNA pair in question into the two different cell strains, and run the experiment. Thus, our measurement kit works best as a preliminary test to get a solid frame of reference for how a synthetase/tRNA pair works in a cell. This application could greatly speed up the process of developing a new synthetase/tRNA pair, as well as characterizing large groups of synthetase/tRNA pairs. | Our ncAA measurement kit was able to successfully compare the fidelity and efficiency of seven different synthetase/tRNA pairs. We were able to confidently say that one synthetase/tRNA pair had a higher fidelity than another. While the measurements may not be perfectly accurate due to the nature of this test, the measurement kit is designed more around ease of use, cost-efficiency, and portability. The ncAA measurement kit can be used to quickly test a synthetase/tRNA pair's quality with minimal effort, as opposed to other more accurate but more intensive or expensive measures such as mass spectrometry. While mass spectrometry can give much more definitive information about a synthetase/tRNA pair, there are many different factors that could make it unreasonable for undergraduate research. It is a technique that requires a fair amount of skill to do properly, and can be very costly as well. For the ncAA measurement kit, all that is necessary is to order the kit parts (pFRYC and PFRY in separate strains of amberless ''E. coli'') from the iGEM registry, transform the synthetase/tRNA pair in question into the two different cell strains, and run the experiment. Thus, our measurement kit works best as a preliminary test to get a solid frame of reference for how a synthetase/tRNA pair works in a cell. This application could greatly speed up the process of developing a new synthetase/tRNA pair, as well as characterizing large groups of synthetase/tRNA pairs. | ||
| Line 229: | Line 228: | ||
Due to the nature of the ncAA Kit Test, <i>in vivo</i> influences will affect certain data and results. This knowledge is very useful by providing a broader and biological perspective of <i>in vivo</i> perspective of cellular health. In general, the presence of ncAAs slowed the growth of amberless ''E. coli'', sometimes quite significantly. This artificially inflated some of the final values that we calculated, which decreased the accuracy of the results. Influences that resulted from ncAA addition such as light-sensitivity, oxidation, and interference with fluorescence readings were accounted for in order to make the final measurements as accurate as possible. | Due to the nature of the ncAA Kit Test, <i>in vivo</i> influences will affect certain data and results. This knowledge is very useful by providing a broader and biological perspective of <i>in vivo</i> perspective of cellular health. In general, the presence of ncAAs slowed the growth of amberless ''E. coli'', sometimes quite significantly. This artificially inflated some of the final values that we calculated, which decreased the accuracy of the results. Influences that resulted from ncAA addition such as light-sensitivity, oxidation, and interference with fluorescence readings were accounted for in order to make the final measurements as accurate as possible. | ||
| + | |||
| + | '''What's your most important result?''' DISCUSS THAT FINDING FIRST. | ||
Are these three paragraphs discussing the results/significance of the results? Yes, we want to talk about possible sources of error, but we don't need three paragraphs for that. We want to talk about what our results mean in a larger context: Does the kit work? Can it be easily improved on? What does our kit do for other labs, for other iGEM teams? For the field of ncAAs? Is the kit definitive by itself? Were there limitations? What might influence good/bad results? Can we unequivocally argue that dopa/3-amino and other ncAA synthetases are worthless? | Are these three paragraphs discussing the results/significance of the results? Yes, we want to talk about possible sources of error, but we don't need three paragraphs for that. We want to talk about what our results mean in a larger context: Does the kit work? Can it be easily improved on? What does our kit do for other labs, for other iGEM teams? For the field of ncAAs? Is the kit definitive by itself? Were there limitations? What might influence good/bad results? Can we unequivocally argue that dopa/3-amino and other ncAA synthetases are worthless? | ||
| Line 240: | Line 241: | ||
<h1>Conclusion</h1> | <h1>Conclusion</h1> | ||
| - | |||
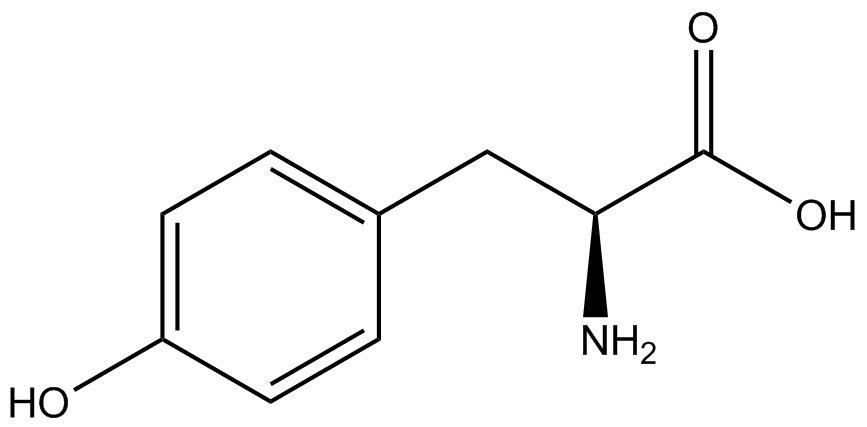
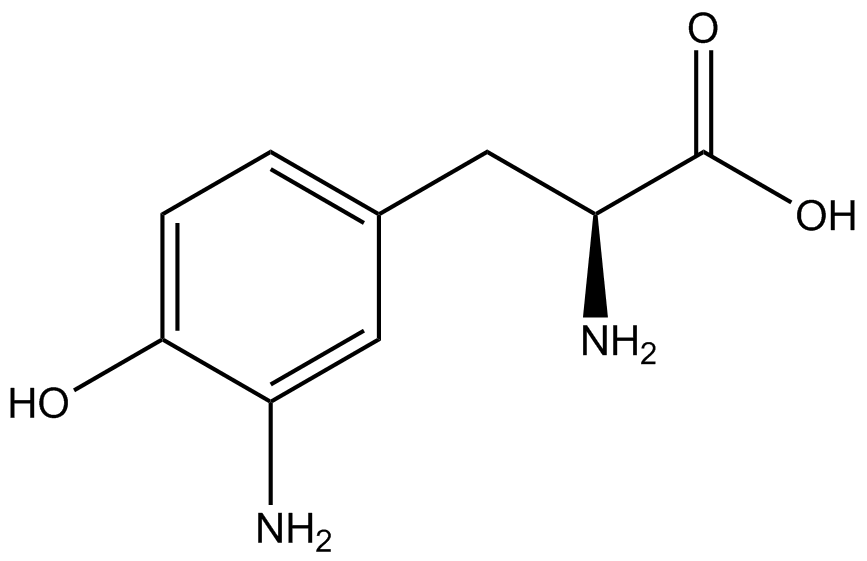
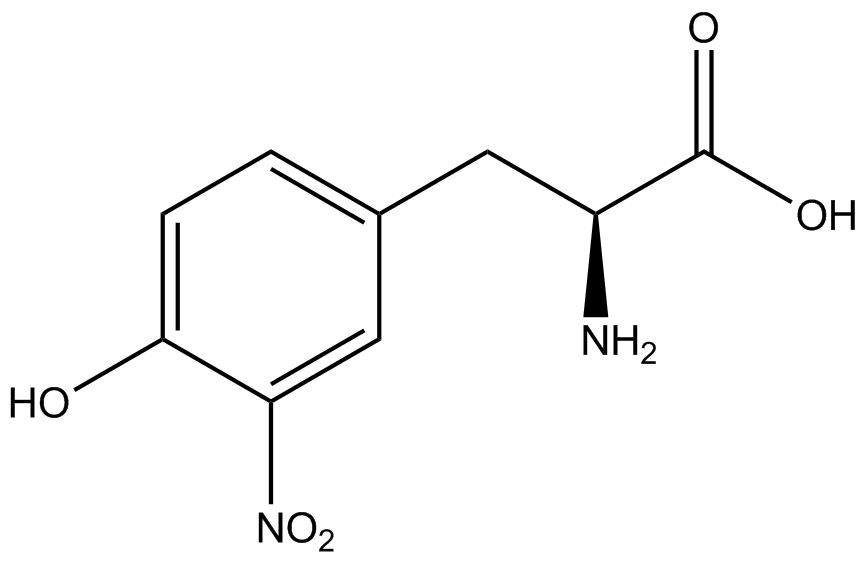
This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | This year the UT Austin iGEM Team has chosen to take part in the [https://2014.igem.org/Tracks/Measurement Measurement Track] of the iGEM competition by contributing to ncAA methods of analysis and fulfilling [https://2014.igem.org/Team:Austin_Texas/medal_requirements medal criteria]. Our team has successfully designed a unique testing kit for ncAA incorporation <i>in vivo</i> using common reporter parts to characterize very uncommon synthetase/tRNA machinery. In addition, we have used our kit to analyze and characterize seven different ncAA synthetase/tRNA pairs as well as tyrosine RS. This method of testing was designed to be undergraduate administrable, transportable, and more affordable than other methods of characterization and analysis. | ||
Revision as of 03:34, 17 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"