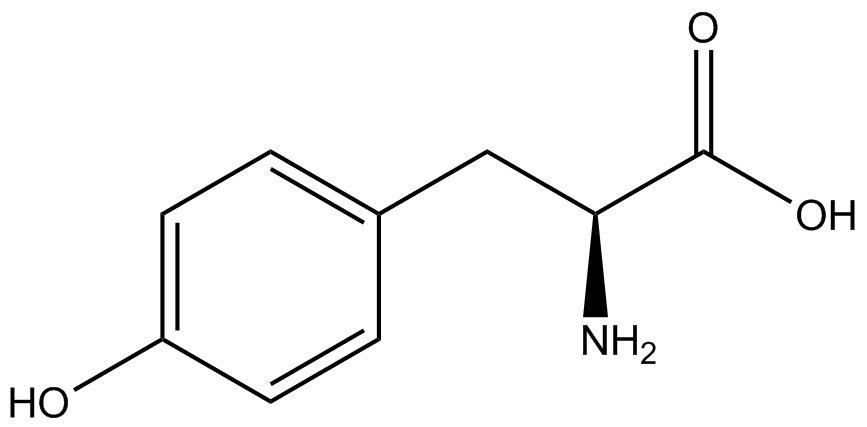
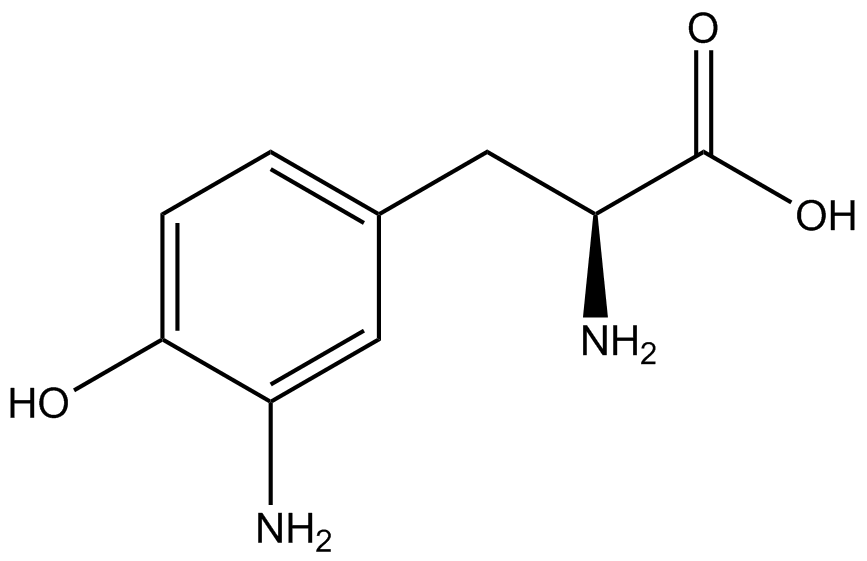
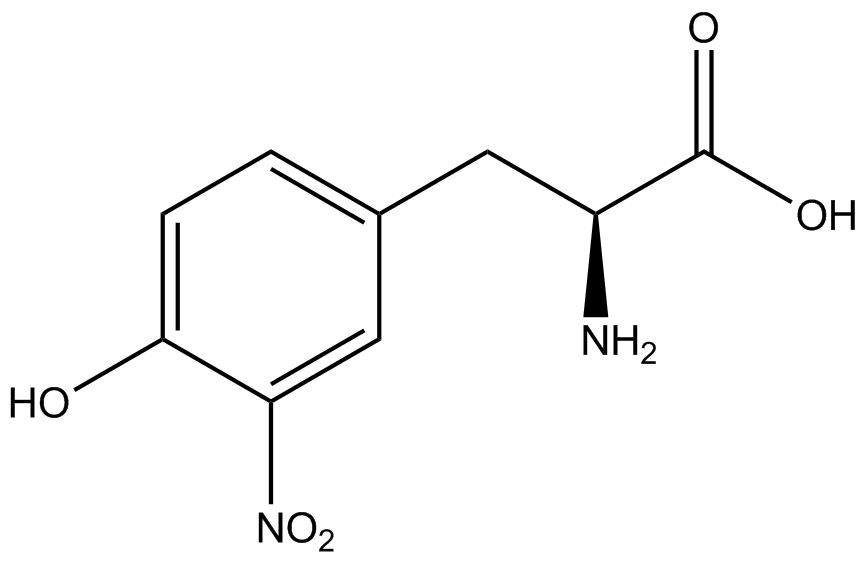
Team:Austin Texas/kit
From 2014.igem.org
(→Plasmids) |
(→Using The Kit) |
||
| Line 123: | Line 123: | ||
To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | To use the kit properly, each culture must have two of the three plasmids from above. There must always be a pStG plasmid as well as either pFRYC or pFRY. However, additional controls can be conducted using only one of the three plasmids. | ||
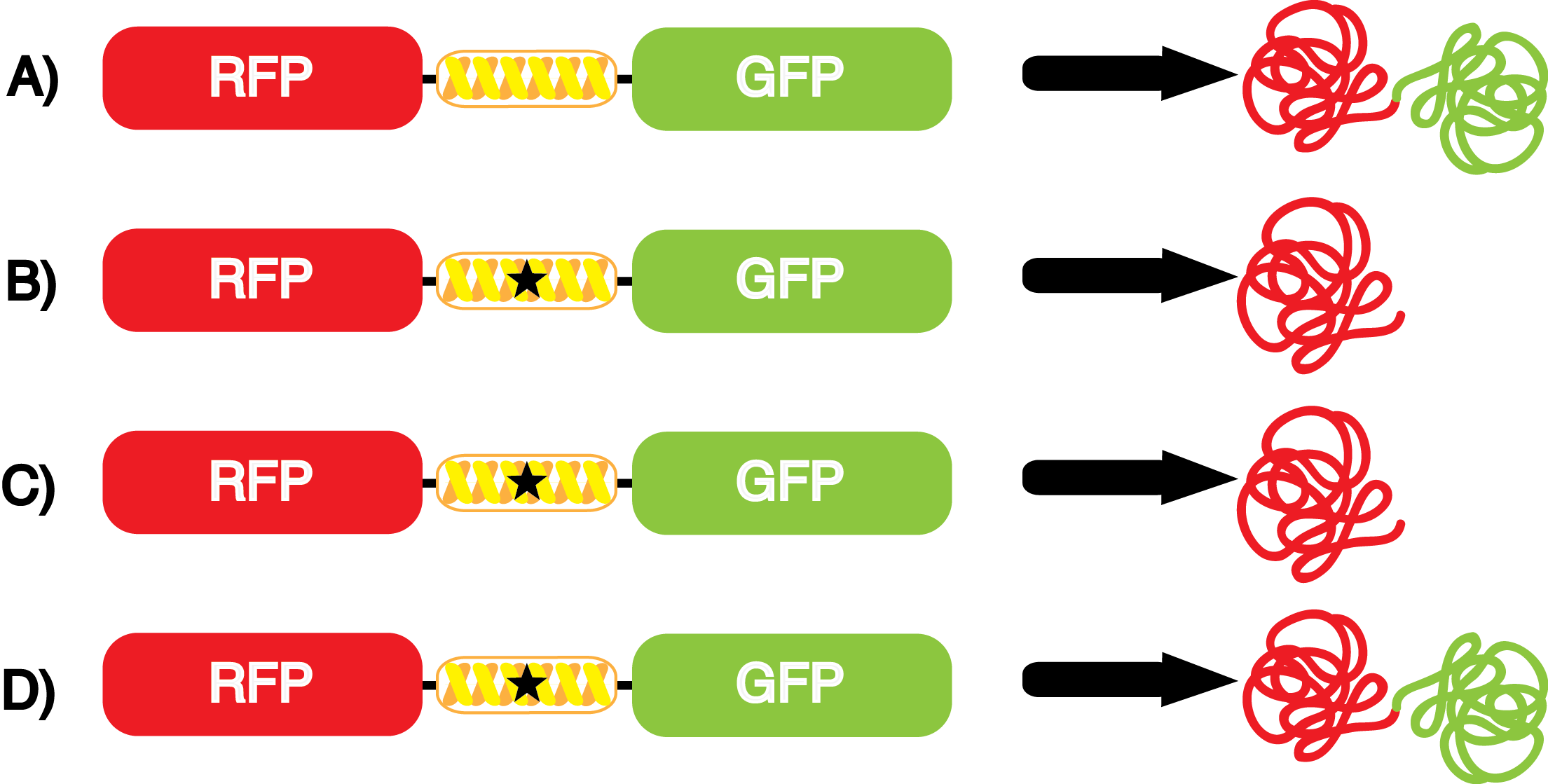
| - | [[File:NcAA kit incorpotation 10-14-14.png|450px|thumb|left|Figure | + | [[File:NcAA kit incorpotation 10-14-14.png|450px|thumb|left|<b>Figure 3</b> Schematic demonstrating the gene expression of the kit plasmids under different growth conditions in the presence of IPTG.<br> |
A) pFRYC (+/-) pStG, (+/-) ncAA.<br> | A) pFRYC (+/-) pStG, (+/-) ncAA.<br> | ||
B) pFRY (-) pStG, (+/-) ncAA.<br> | B) pFRY (-) pStG, (+/-) ncAA.<br> | ||
| Line 130: | Line 130: | ||
The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient at conducting every step in the process of reading through an amber codon.]] | The above results represent the "perfect" tRNA synthetase/tRNA pair: one that has 100% fidelity and 100% efficiency. Actual synthetase/tRNA pairs will occasionally misincorporate an incorrect amino acid, and these pairs will not always be perfectly efficient at conducting every step in the process of reading through an amber codon.]] | ||
| - | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence (Figure | + | *''' A)''' In a cell containing pStG and pFRYC, the ribosome will translate the RFP reporter, linker, and sfGFP, producing red and green fluorescent proteins that result in visible yellow fluorescence (Figure 3A). This happens because the reporter contains no amber codon, and thus does not require a ncAA synthetase/tRNA pair. |
| - | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence (Figure | + | *''' B)''' If a cell contained only pFRY, without pStG, then the ribosome will translate the RFP and terminate at the amber codon on the linker producing a red fluorescence (Figure 3B). This occurs because without the pStG, the amber codon is not recoded to allow for ncAA incorporation. |
| - | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence (Figure | + | * '''C)''' In a cell containing pStG and pFRY, there are multiple possible outcomes, depending on what is present in the culture. In the absence of ncAA, the ribosome will translate the RFP and terminate at the amber stop codon on the linker producing a red fluorescence (Figure 3C). This occurs because while the codon has been recoded, there is no ncAA and without ncAA there can be no incorporation at the amber codon. |
| - | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence (Figure | + | *''' D)''' In a cell containing pStG and pFRY, there are multiple possible outcomes. In the PRESENCE of ncAA, the ribosome will translate the RFP, then it incorporates the ncAA at the amber codon in the linker, and finally it proceeds to translate the downstream sfGFP reporter, producing a visible yellow fluorescence (Figure 3D). This occurs because the synthetase found in pStG can covalently attach the ncAA to the tRNA gene present in pStG. This "charged tRNA" is then present during translation, allowing incorporation of the ncAA into the reporter protein. |
However, the above scenarios assume that the synthetase/tRNA pair function efficiently and with high fidelity. | However, the above scenarios assume that the synthetase/tRNA pair function efficiently and with high fidelity. | ||
| - | *Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. This "misincorporation" means that such a synthetase/tRNA pair would not give results that mimic Figure | + | *Actual synthetase/tRNA pairs sometimes have '''low fidelity''', leading to the incorporation of an incorrect amino acid. This "misincorporation" means that such a synthetase/tRNA pair would not give results that mimic Figure 3C in the absence of ncAA. Rather, in the absence of ncAA, such a pair would give results similar to Figure 3D, full expression of RFP and sfGFP. High rates of misincorporation are equivalent to '''low fidelity''', and tRNA synthetases with low fidelity are not ideal. |
| - | *Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure | + | *Actual synthetase/tRNA pairs can also sometimes have '''low efficiency'''. As these are artificially selected pairs, sometimes they do not function as well as natural synthetase/tRNA pairs. In such a case, the amount of sfGFP translated in Figure 3D might be significantly lower than expected. '''low efficiency''' is also not ideal as it may limit levels of protein expression. |
==Experimental Preparation== | ==Experimental Preparation== | ||
Revision as of 02:48, 17 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"