Team:Austin Texas/kit
From 2014.igem.org
(→Motivation) |
(→Fidelity of Incorporation) |
||
| Line 194: | Line 194: | ||
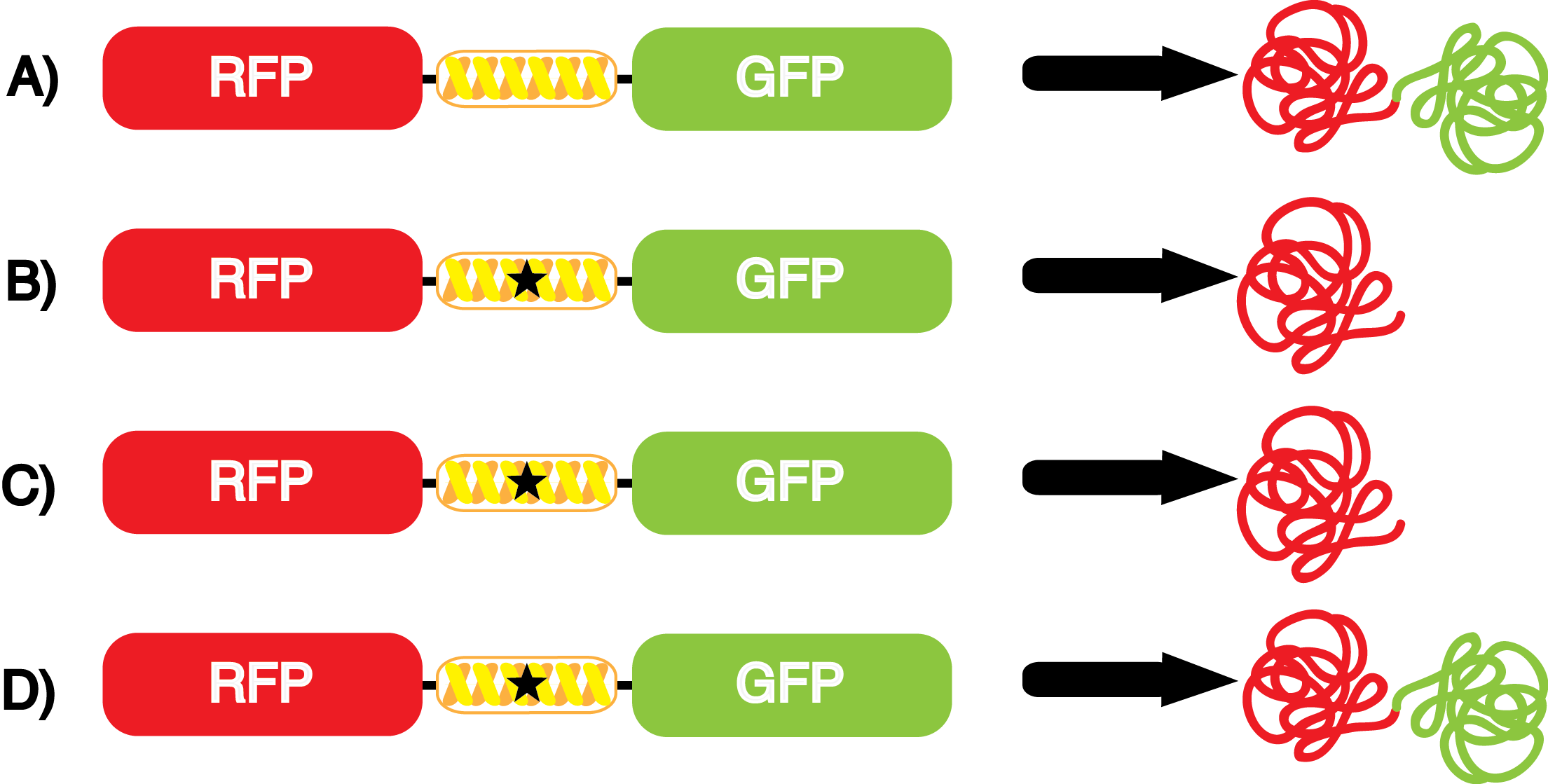
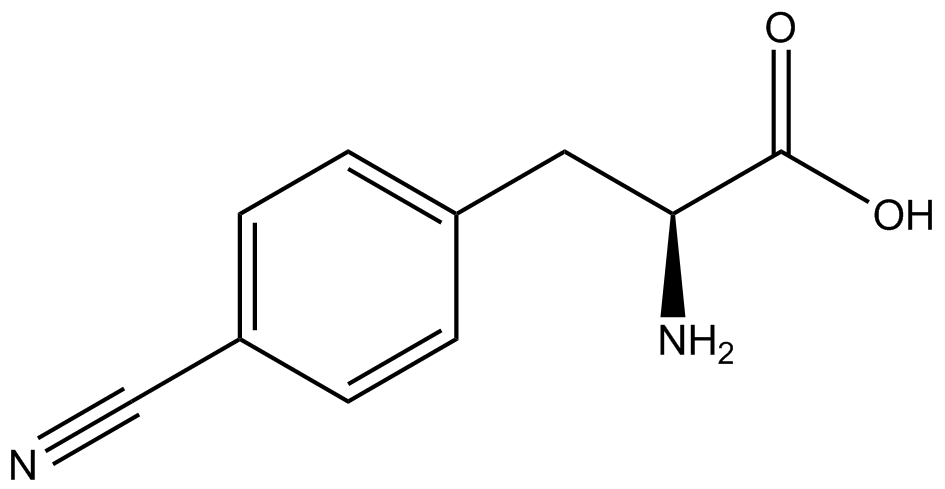
''italics = methods Strains containing pFRY and pStG plasmids were grown in the presence and absence of the corresponding non-canonical amino acid. The fluorescence of RFP and GFP readings of each culture were recorded at a culture density of 0.5 OD 600.'' | ''italics = methods Strains containing pFRY and pStG plasmids were grown in the presence and absence of the corresponding non-canonical amino acid. The fluorescence of RFP and GFP readings of each culture were recorded at a culture density of 0.5 OD 600.'' | ||
| - | + | ||
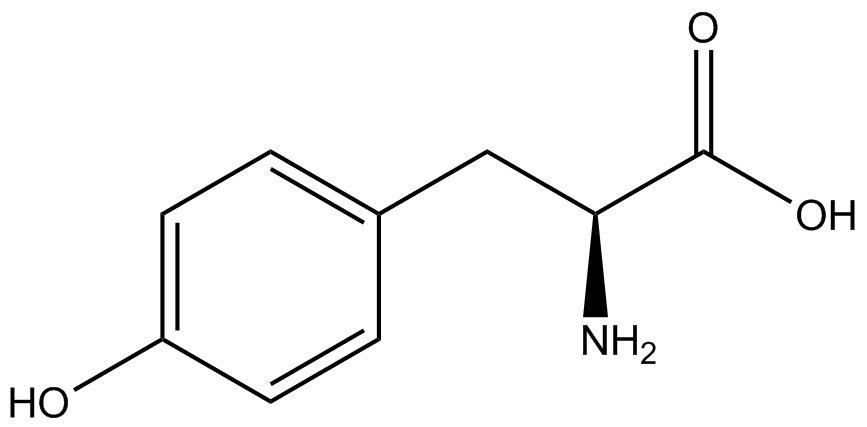
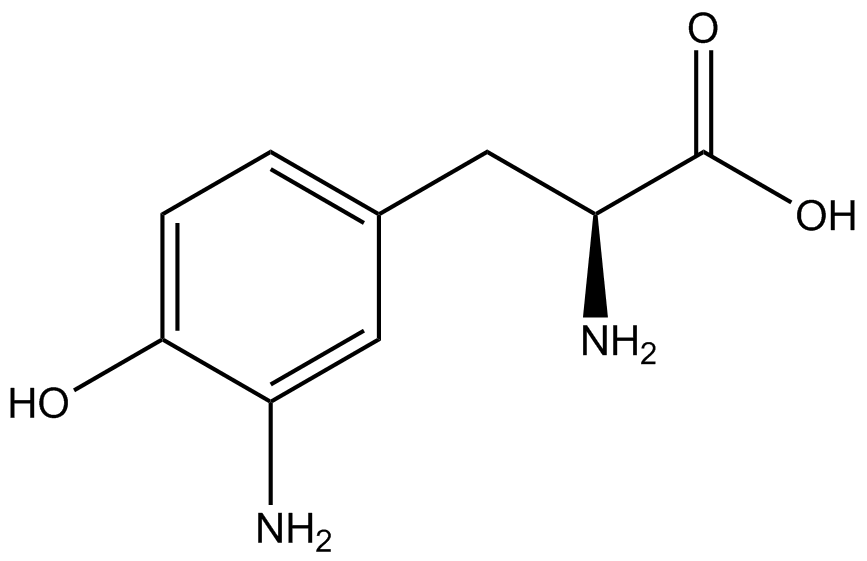
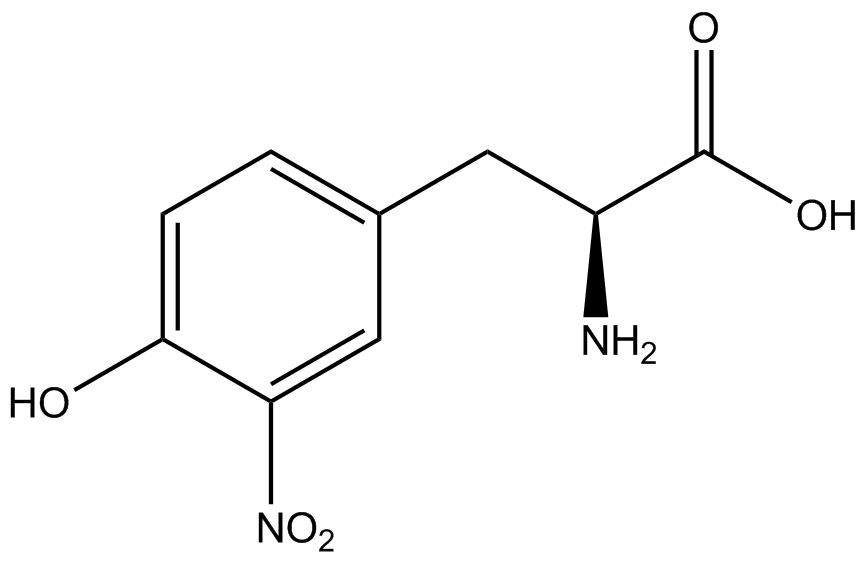
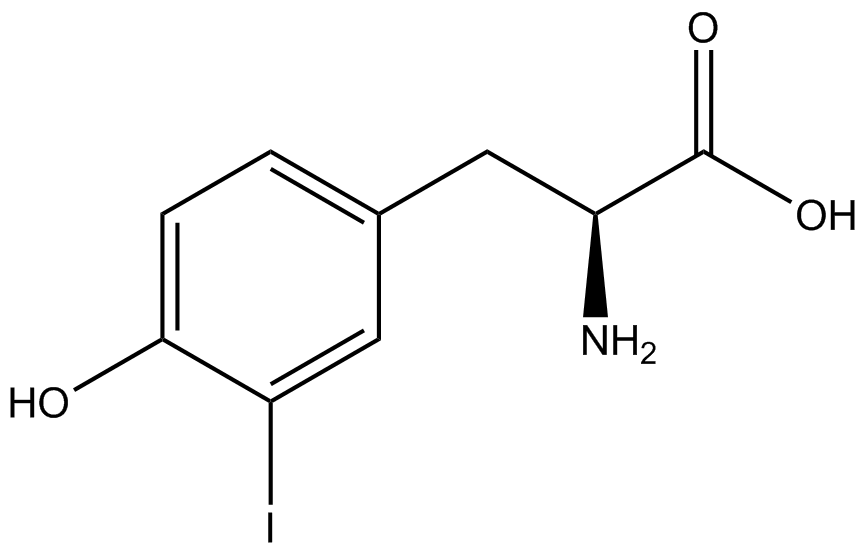
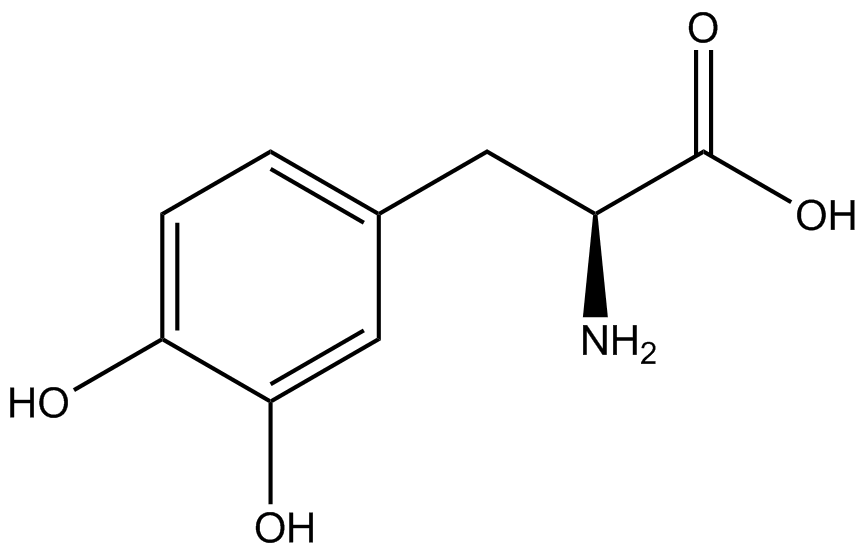
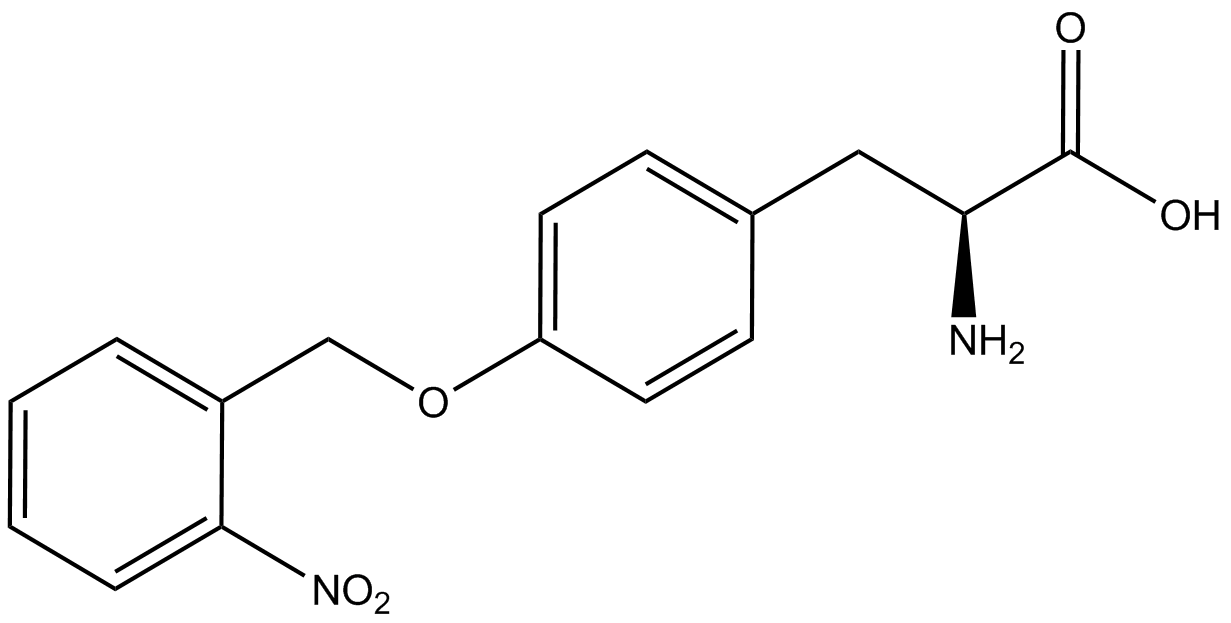
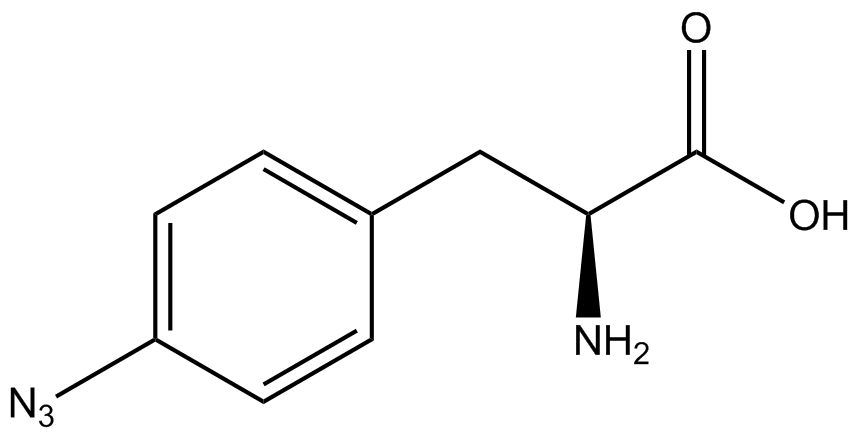
| - | GFP | + | To determine the change in GFP fluorescence when the ncAA was present, we first had to calculate how much GFP was expressed relative to the RFP, which would give an upper estimate of how much GFP could theoretically be expressed. We first divided both the GFP and RFP levels by the OD 600 of the culture in order to get the per cell fluorescence levels. We then normalized the GFP fluorescence for one culture to its RFP fluorescence so that we could compare the GFP fluorescence levels between cultures. The normalized GFP values were then compared between cultures grown in the presence of ncAA and cultures grown in the absence of ncAA, which would indicate how the level of GFP fluorescence changes when the ncAA is present. When these values were graphed (Figure 3), some synthetase/tRNA pairs such as 4-azidophenylalanine, 3-nitrotyrosine, 3-iodotyrosine, and ortho-nitrobenzyltyrosine resulted in higher GFP fluorescence in the presence of ncAA than in the absence of ncAA, which suggests that those synthetases only incorporated an amino acid if their specific amino acid was present, meaning that they have a high fidelity. However, the other synthetase/tRNA pairs (3-aminotyrosine, L-DOPA, and cyanophenylalanine) did not show a significant increase in GFP fluorescence normalized to RFP fluorescence when the ncAA was present, indicating that they would incorporate other amino acids at the amber stop codon when their specific amino acid was not present (and perhaps even when it was), and thus have a low fidelity. |
| - | + | ||
| - | + | ||
==Synthetase efficiency== | ==Synthetase efficiency== | ||
Revision as of 00:49, 17 October 2014
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"