Team:Austin Texas/kit
From 2014.igem.org
(→Discussion) |
(→Results and Data) |
||
| Line 161: | Line 161: | ||
==Accuracy of Incorporation== | ==Accuracy of Incorporation== | ||
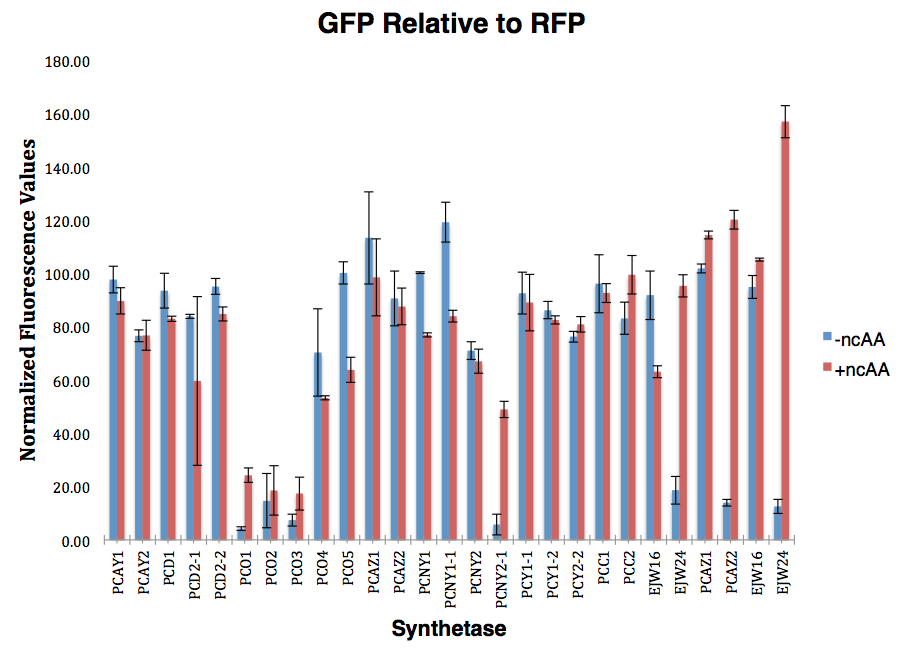
| - | [[File:10-14-14 (screenshot) GFP Relative to RFP.png|thumb|600px|Figure | + | [[File:10-14-14 (screenshot) GFP Relative to RFP.png|thumb|600px|Figure 3. Graph showing the level of GFP fluorescence relative to RFP fluorescence.]] |
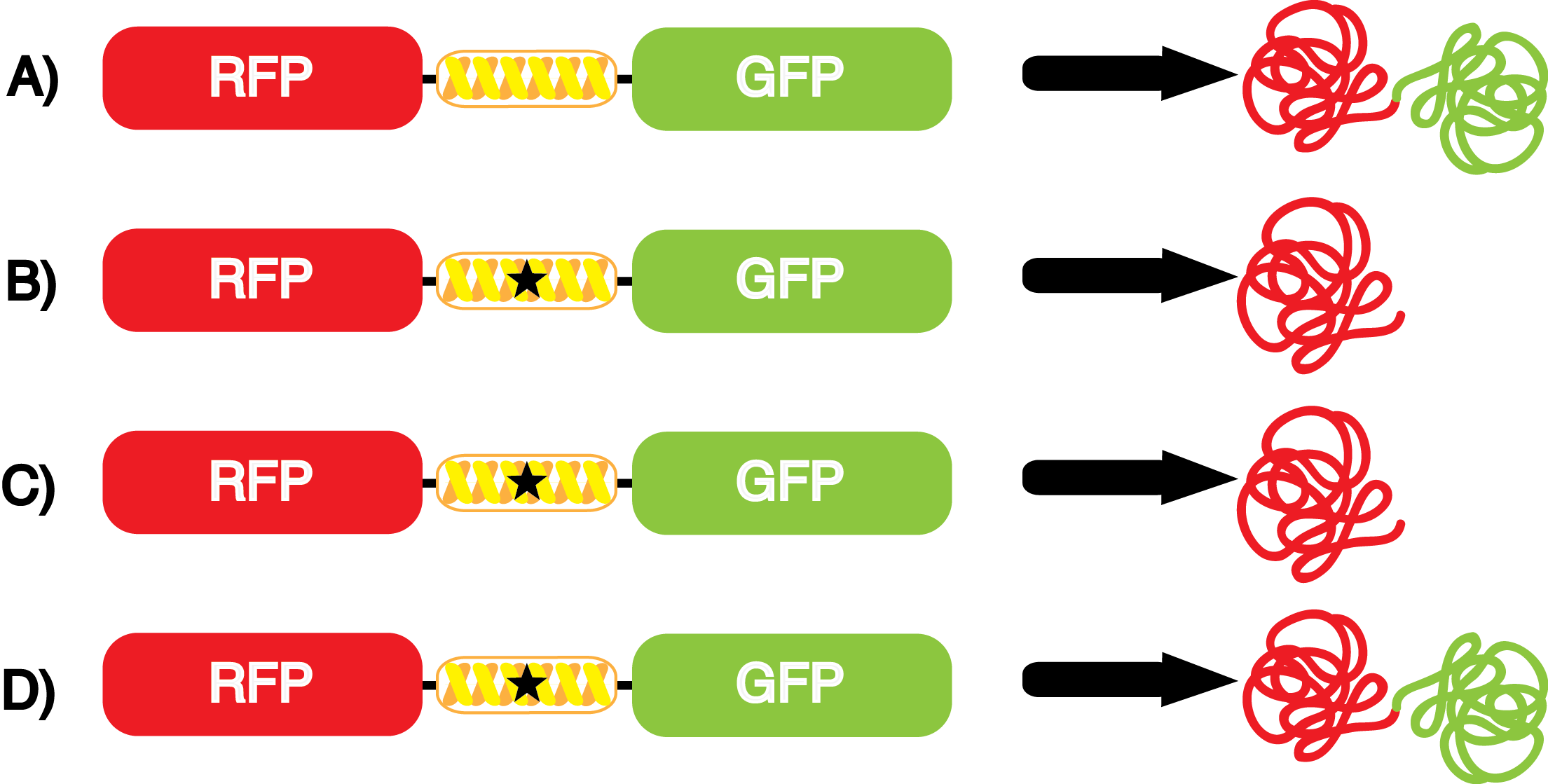
The accuracy of each ncAA sythetase/tRNA pair was measured by comparing the GFP production of pStG/pFRY strains in (+/-) ncAA conditions. In the absence of a non-cannonical only RFP should be translated. Translation is expected to terminate between RFP and GFP at the amber stop codon (UAG) on linker sequence on pFRY. Alternatively, translation should continue through the linker in the presence of a certain non-cannonical due to its incorporation at UAG. In this case, RFP and GFP should both be translated. The fluorescence of RFP and GFP is detectable with fluorometer. | The accuracy of each ncAA sythetase/tRNA pair was measured by comparing the GFP production of pStG/pFRY strains in (+/-) ncAA conditions. In the absence of a non-cannonical only RFP should be translated. Translation is expected to terminate between RFP and GFP at the amber stop codon (UAG) on linker sequence on pFRY. Alternatively, translation should continue through the linker in the presence of a certain non-cannonical due to its incorporation at UAG. In this case, RFP and GFP should both be translated. The fluorescence of RFP and GFP is detectable with fluorometer. | ||
| Line 173: | Line 173: | ||
==Incorporation Value== | ==Incorporation Value== | ||
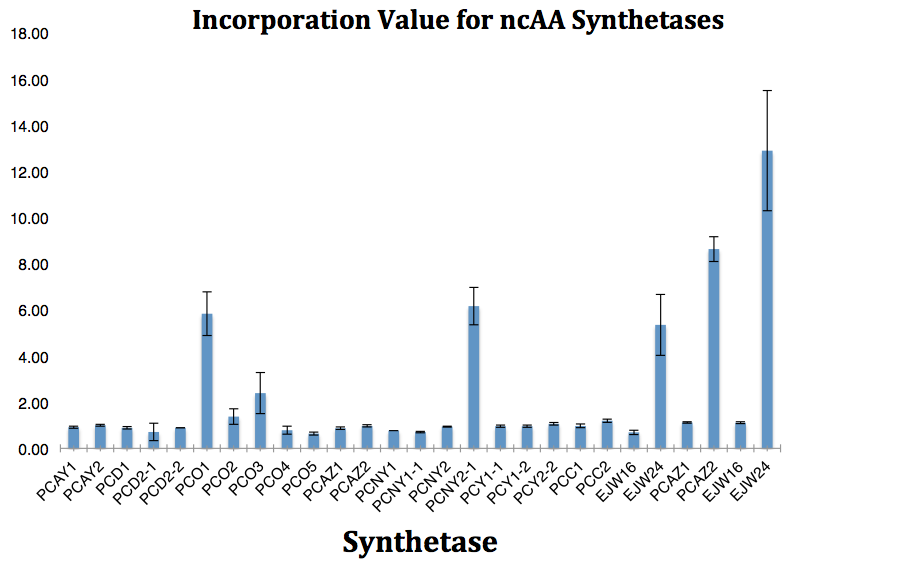
| - | [[File:10-14-14 (snapshot) Incorporation Value for ncAA Synthetasaes.png|600px|thumb|Figure | + | [[File:10-14-14 (snapshot) Incorporation Value for ncAA Synthetasaes.png|600px|thumb|Figure 4.<br> |
<i>May need to consider a different naming convention or the cultures. Possibly AY-C and AY-E for Control and Experimental?? </i> The fluorescence and OD600 readings of each culture were used to calculate a value for incorporation efficiency of each synthetase. <i>Need to add an explanation of how these values were calculated</i>]] | <i>May need to consider a different naming convention or the cultures. Possibly AY-C and AY-E for Control and Experimental?? </i> The fluorescence and OD600 readings of each culture were used to calculate a value for incorporation efficiency of each synthetase. <i>Need to add an explanation of how these values were calculated</i>]] | ||
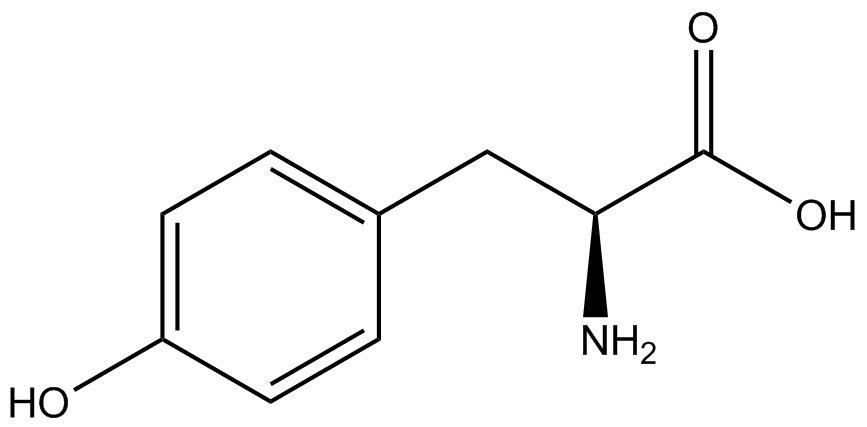
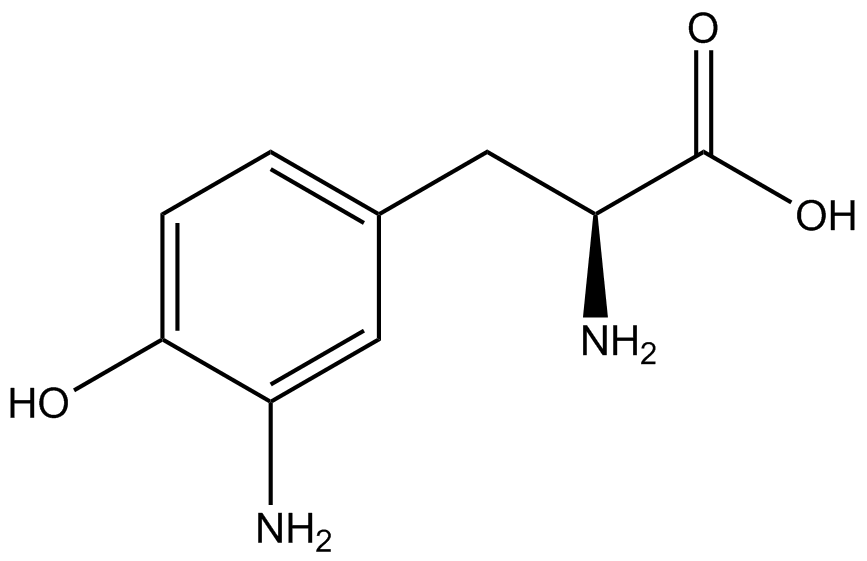
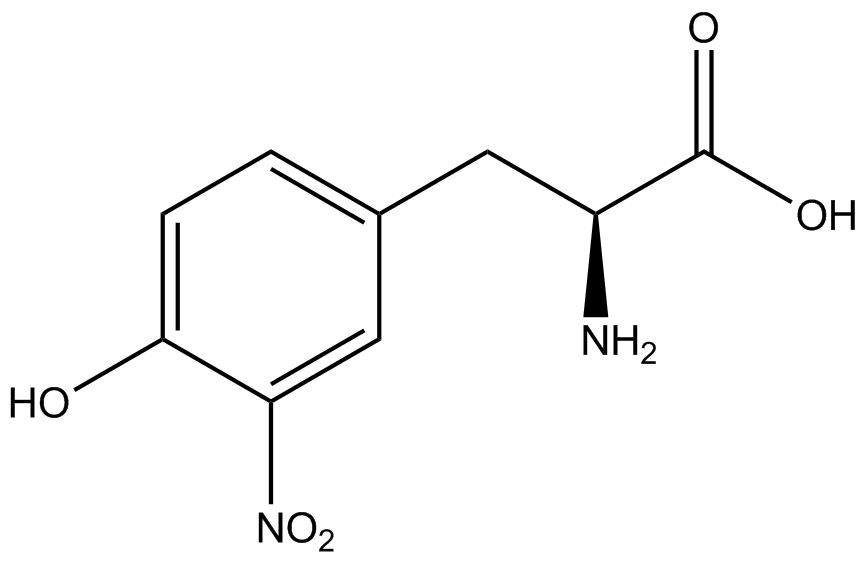
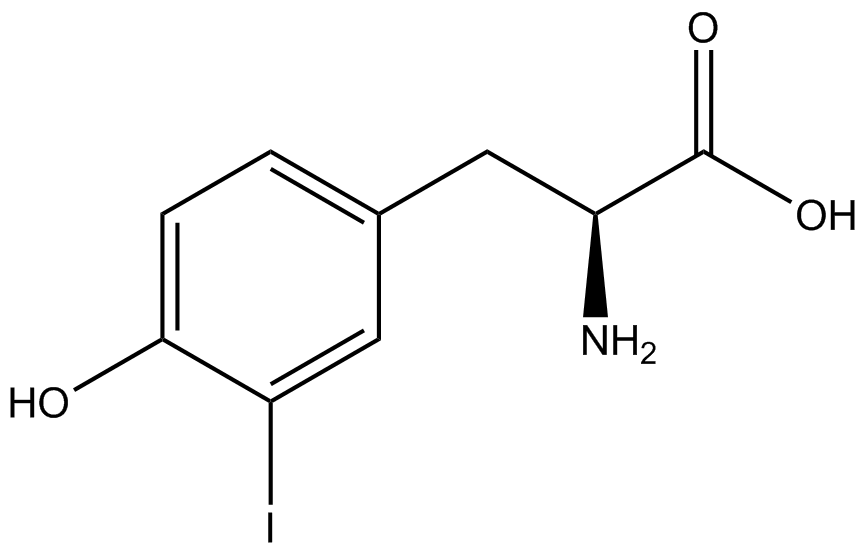
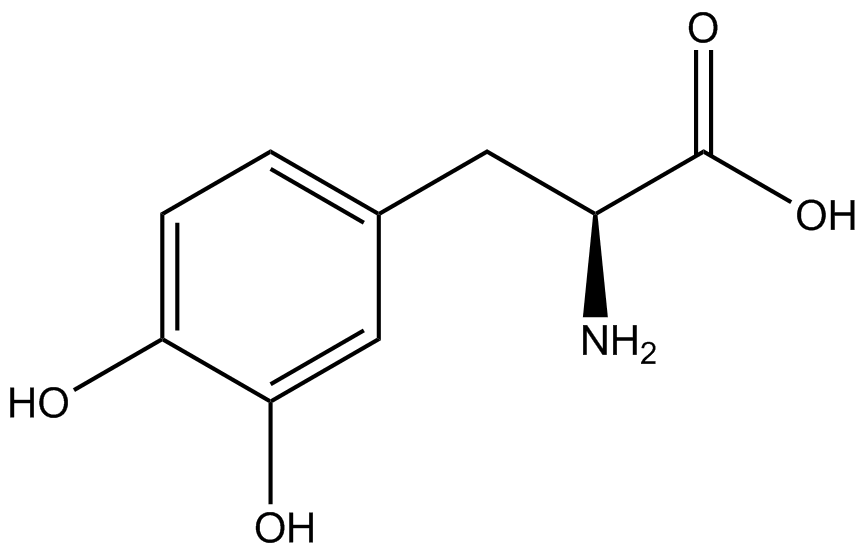
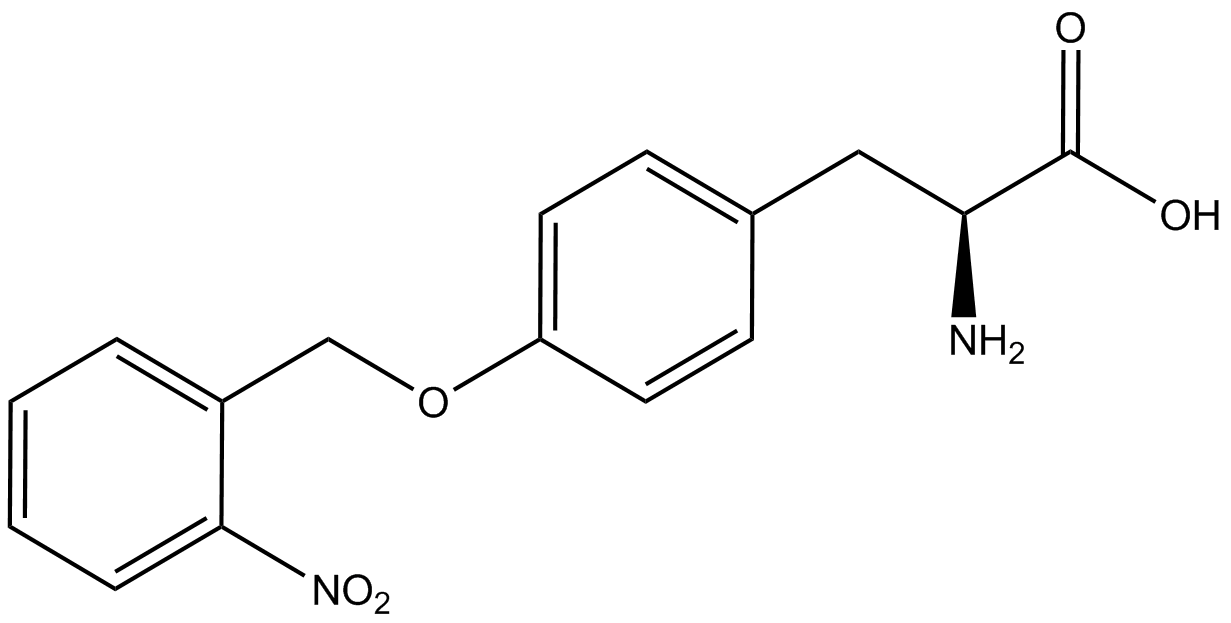
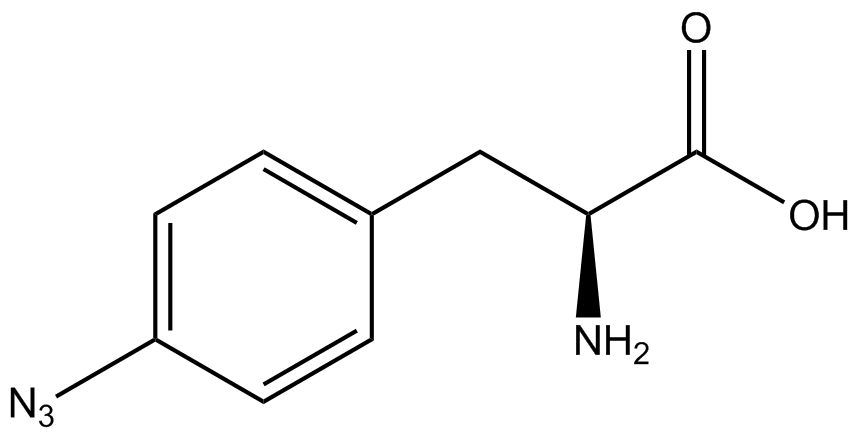
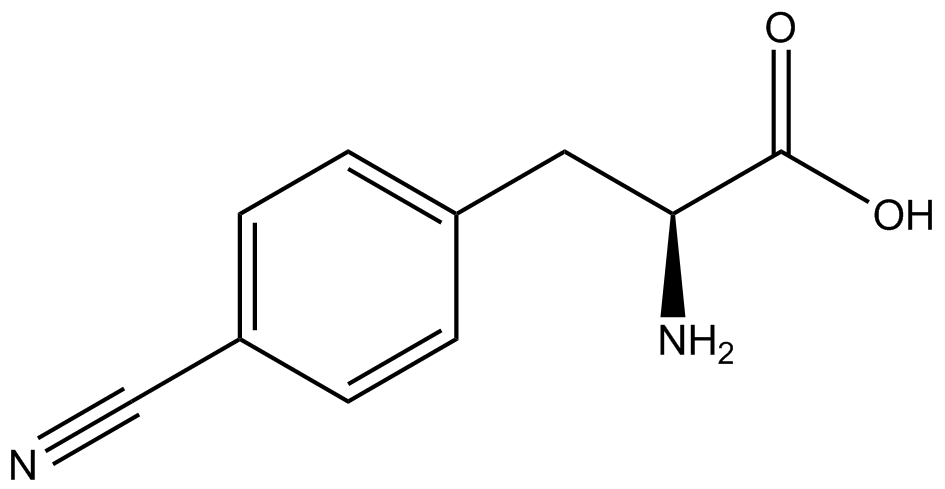
The fluorescence and OD600 readings of each culture were compared to evaluate the level of incorporation by each synthetase/tRNA pair (pStG). The synthetase/tRNA pairs that showed the highest level of ncAA incorporation include 3-iodotyrosine, 4-azidophenylalanine, 3-nitrotyrosine, and ortho-nitrobenzyltyrosine in decreasing order. Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly suggesting possible toxicity to the cell however ncAA incorporation remained high. Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation include 3-aminotyrosine, L-dopa, tyrosine, and 4-cyanophenylalanine. | The fluorescence and OD600 readings of each culture were compared to evaluate the level of incorporation by each synthetase/tRNA pair (pStG). The synthetase/tRNA pairs that showed the highest level of ncAA incorporation include 3-iodotyrosine, 4-azidophenylalanine, 3-nitrotyrosine, and ortho-nitrobenzyltyrosine in decreasing order. Among the ncAAs tested, ONBY consistently slowed the growth rate of the culture significantly suggesting possible toxicity to the cell however ncAA incorporation remained high. Synthetase/tRNA pairs that showed the lowest levels of ncAA incorporation include 3-aminotyrosine, L-dopa, tyrosine, and 4-cyanophenylalanine. | ||
| - | |||
=Discussion= | =Discussion= | ||
Revision as of 23:52, 14 October 2014
|
 "
"