Team:Austin Texas/kit
From 2014.igem.org
Jordanmonk (Talk | contribs) m (→Kit Introduction) |
Jordanmonk (Talk | contribs) m (→Kit Introduction) |
||
| Line 77: | Line 77: | ||
==Kit Introduction== | ==Kit Introduction== | ||
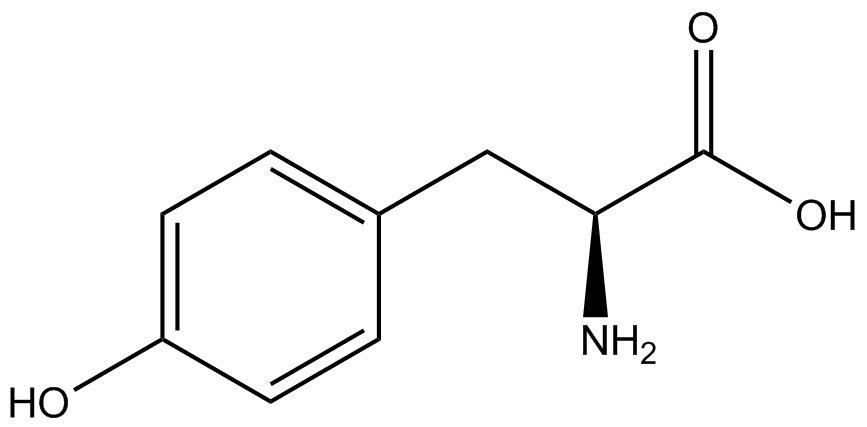
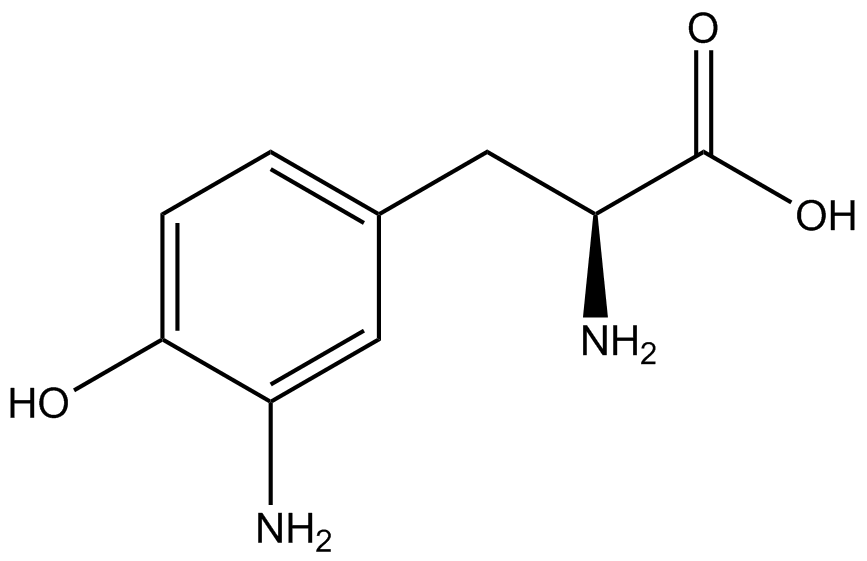
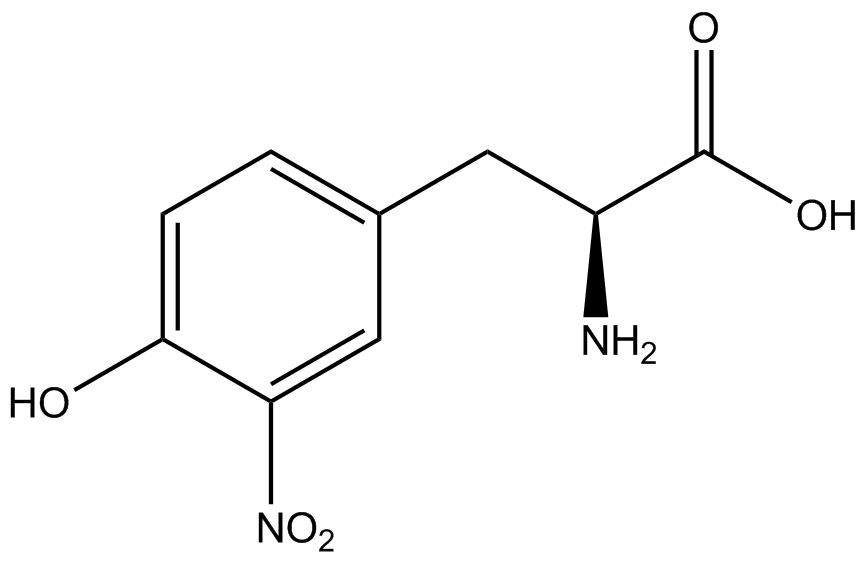
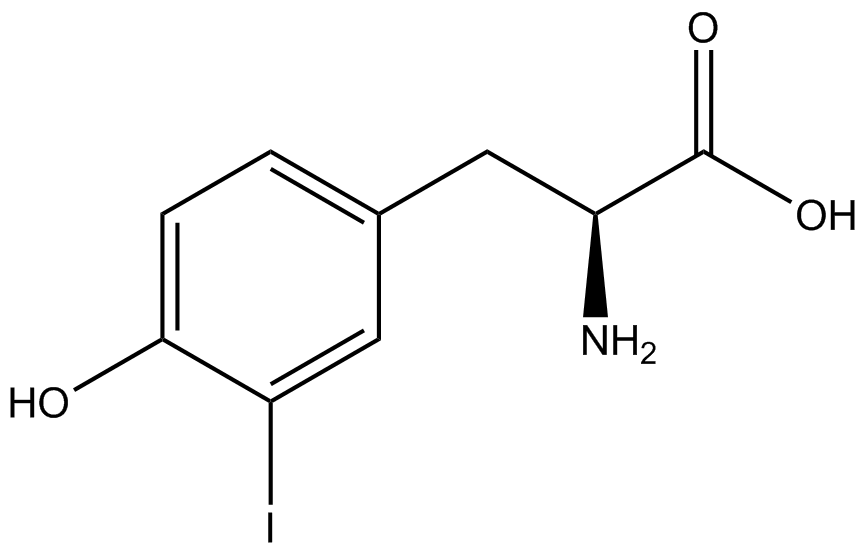
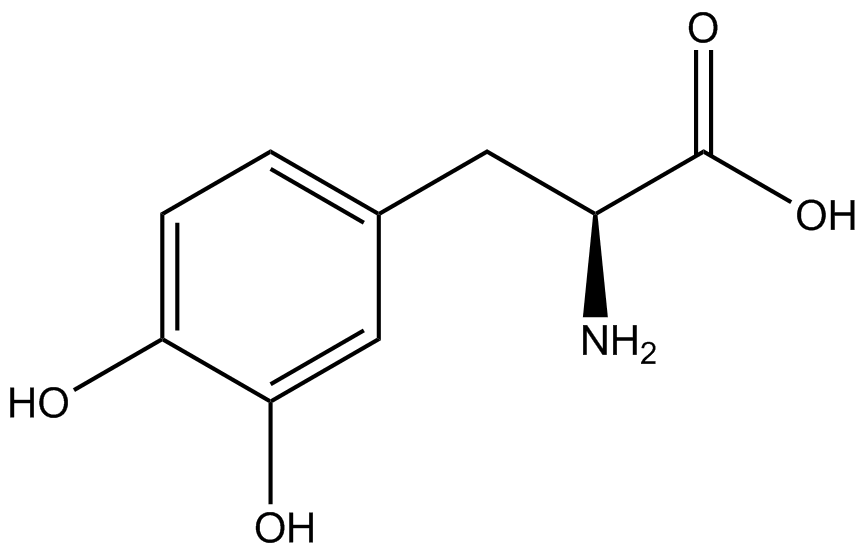
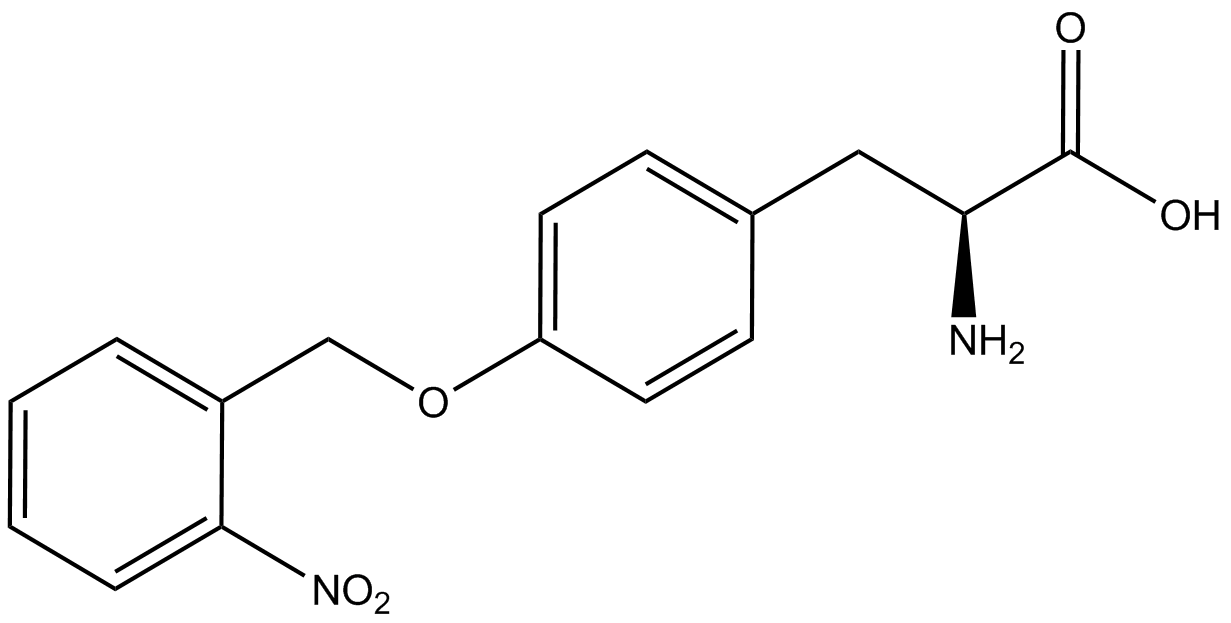
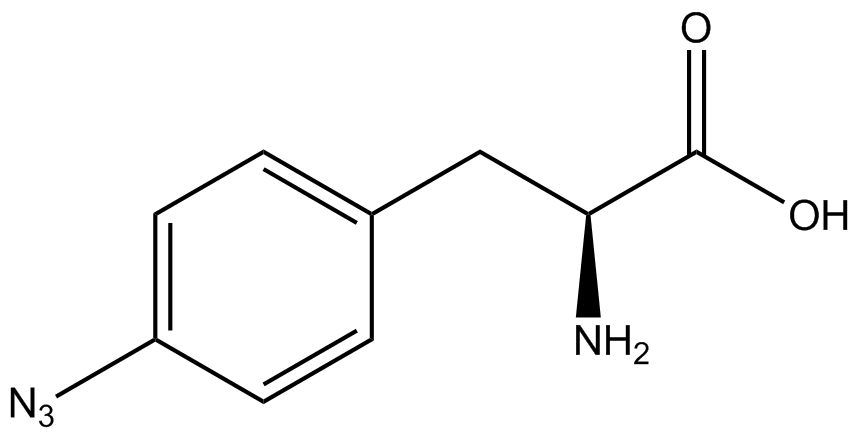
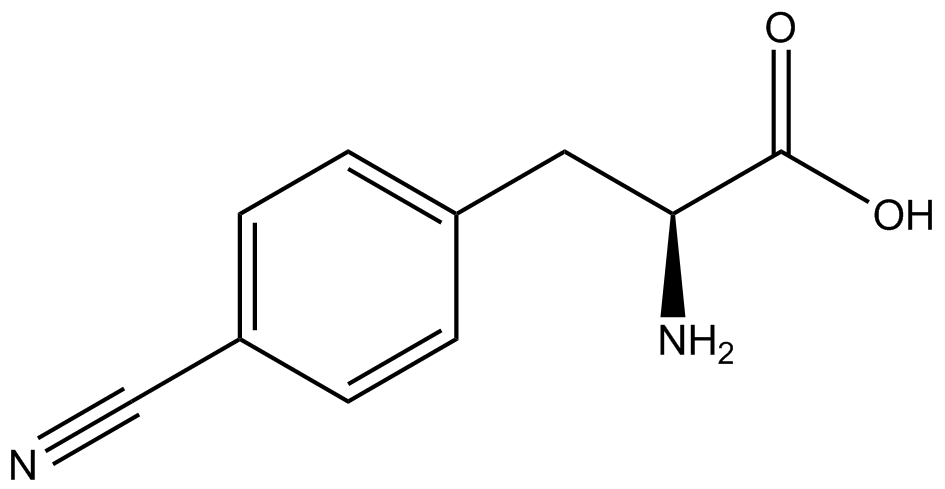
| - | In recent years the ability to expand the genetic code has been made possible by re-coding the amber stop codon UAG via the use of modified tRNA synthetase/tRNA pairs. These synthetase/tRNA pairs act together to charge the tRNA with a non-canonical amino acid (ncAA), an amino acid that is not one of the 20 amino acids normally encoded by a codon. While the library of ncAA synthetase/tRNA pairs continues to grow, the properties of each pairing have yet to be systematically characterized using a standardized methodology. The 2014 University of Texas at Austin iGEM Team has developed a method that allows for efficient in vivo characterization, both qualitative and quantitative, of these novel synthetase/tRNA pairs. | + | In recent years the ability to expand the genetic code has been made possible by re-coding the amber stop codon, UAG, via the use of modified tRNA synthetase/tRNA pairs. These synthetase/tRNA pairs act together to charge the tRNA with a non-canonical amino acid (ncAA), an amino acid that is not one of the 20 amino acids normally encoded by a codon. While the library of ncAA synthetase/tRNA pairs continues to grow, the properties of each pairing have yet to be systematically characterized using a standardized methodology. The 2014 University of Texas at Austin iGEM Team has developed a method that allows for efficient in vivo characterization, both qualitative and quantitative, of these novel synthetase/tRNA pairs. |
'''Desired properties??? High fidelity? High efficiency of incorporation? We should indicate what we want our kit to test for.''' | '''Desired properties??? High fidelity? High efficiency of incorporation? We should indicate what we want our kit to test for.''' | ||
Revision as of 09:10, 14 October 2014
|
 "
"