Team:Austin Texas/kit
From 2014.igem.org
Jordanmonk (Talk | contribs) (→Experimental Design and Method) |
Jordanmonk (Talk | contribs) (→Experimental Design and Method) |
||
| Line 95: | Line 95: | ||
In order to recode UAG, a synthetase must be mutated to effectively grab onto an ncAA. Various methods of directed evolution are typically used to modify a synthetase such that it can accept and charge a non-canonical amino acid. The library of ncAA synthetases available have ranging levels of reported efficiency and are not well characterized. This year the UT iGEM Team created a test kit designed to characterize the efficiency of any ncAA synthetase/tRNA pair. | In order to recode UAG, a synthetase must be mutated to effectively grab onto an ncAA. Various methods of directed evolution are typically used to modify a synthetase such that it can accept and charge a non-canonical amino acid. The library of ncAA synthetases available have ranging levels of reported efficiency and are not well characterized. This year the UT iGEM Team created a test kit designed to characterize the efficiency of any ncAA synthetase/tRNA pair. | ||
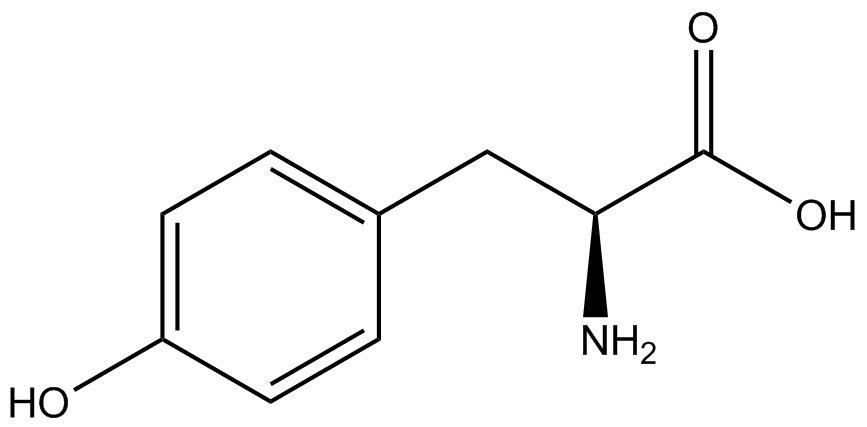
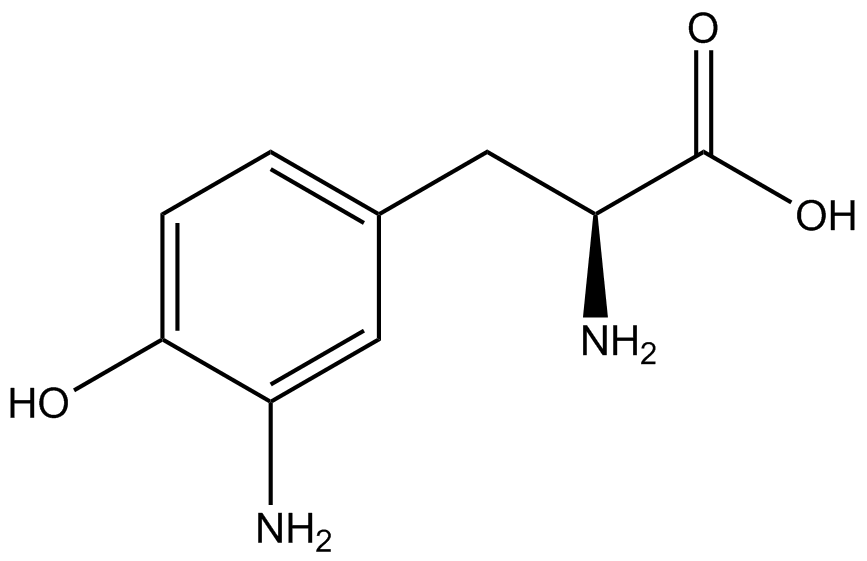
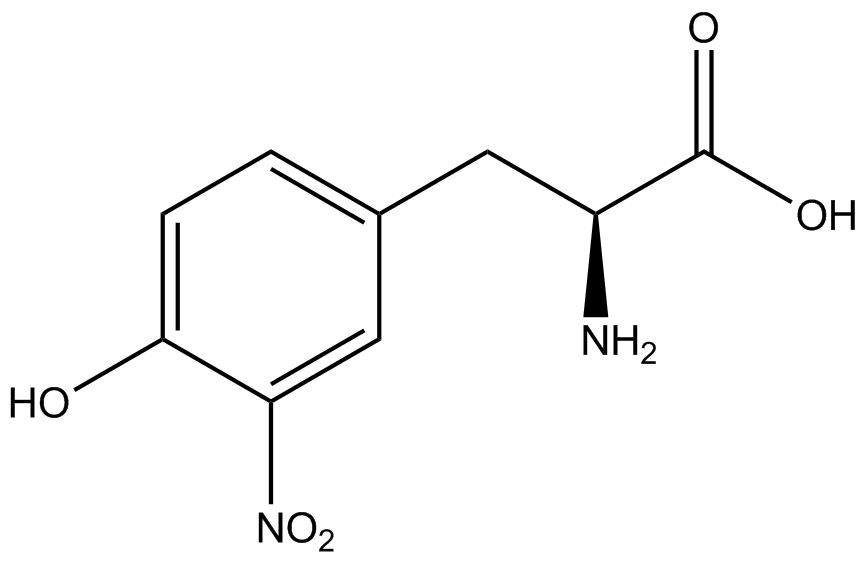
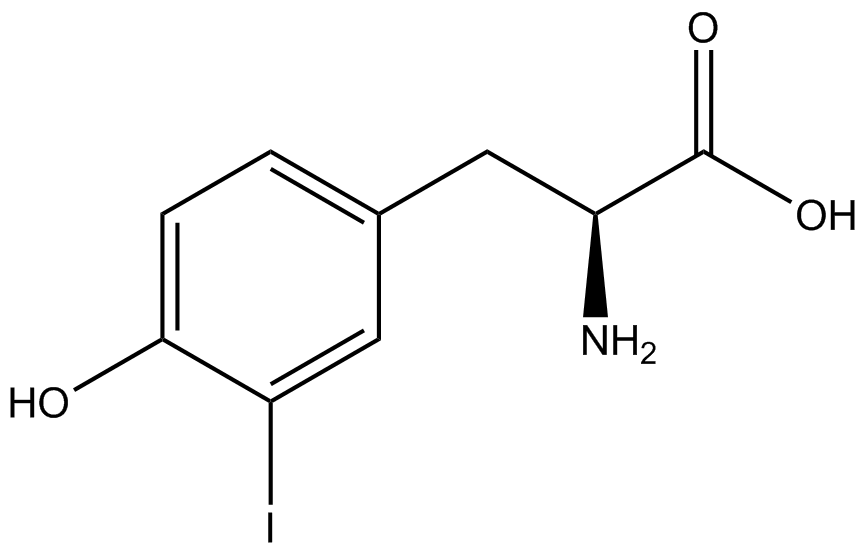
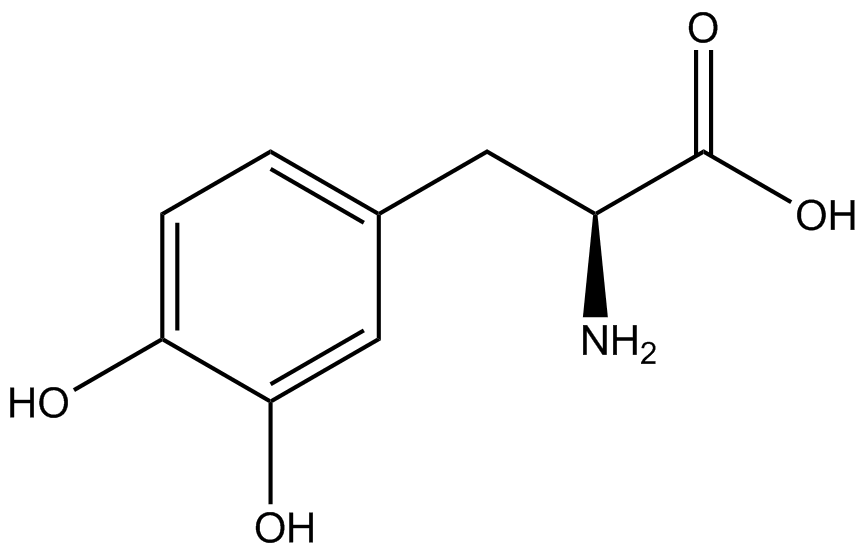
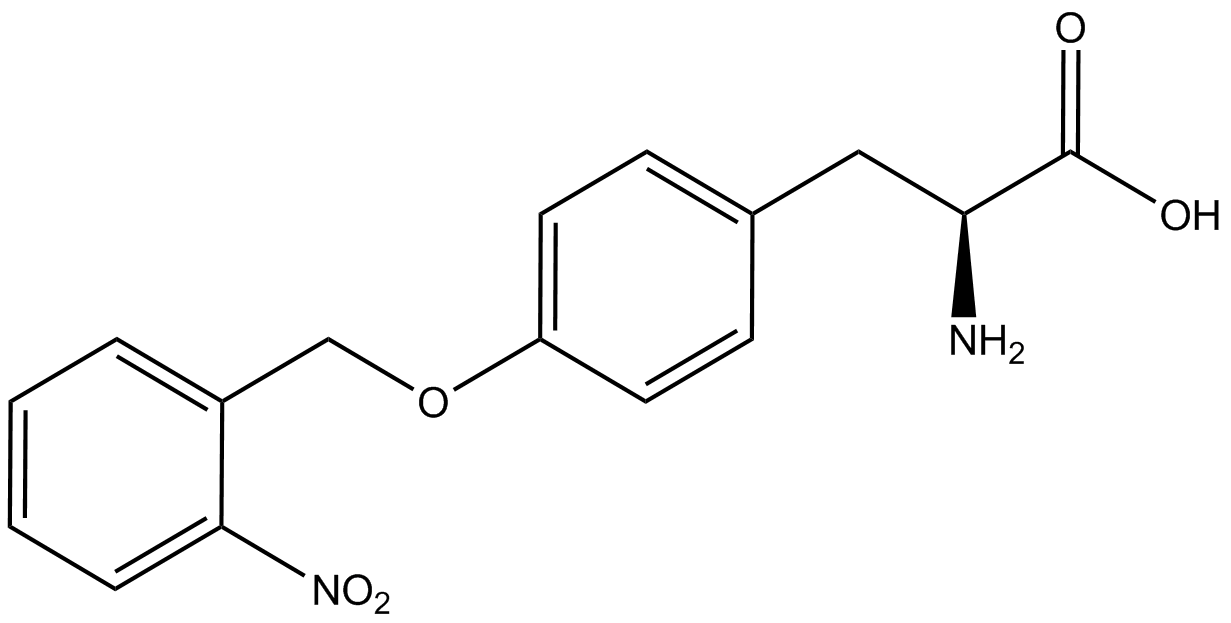
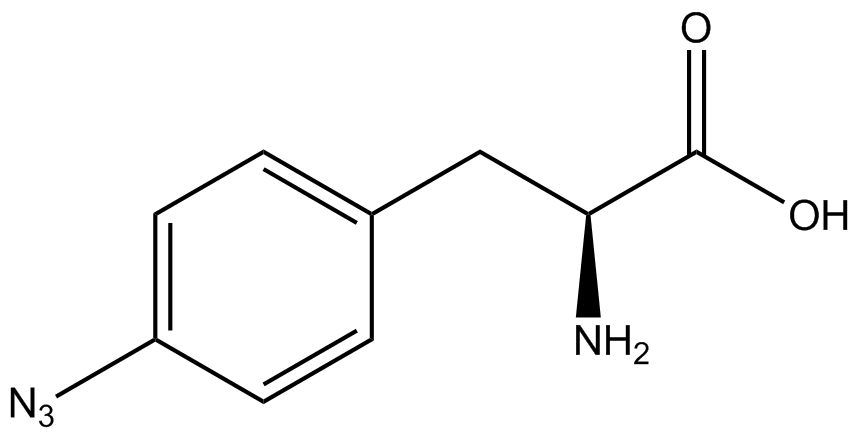
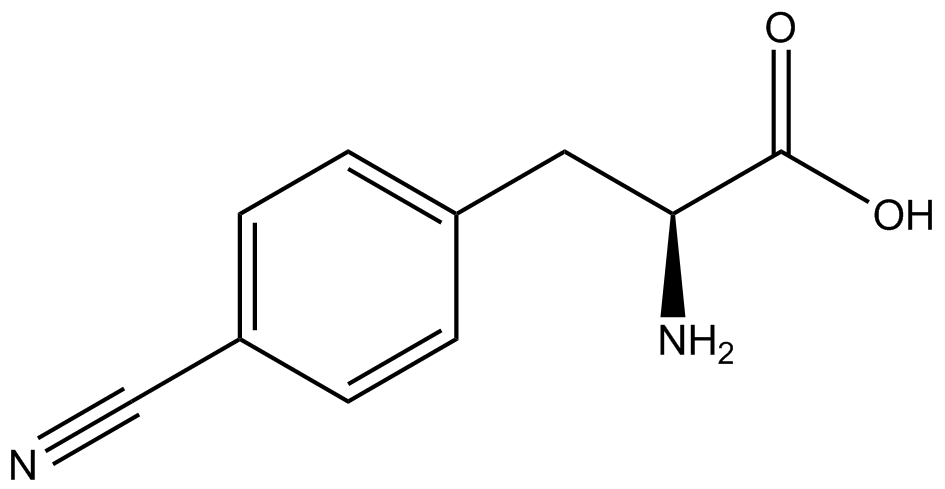
| - | A total of seven | + | A total of seven ncAAs were used in addition to tyrosine. [https://2014.igem.org/Team:Austin_Texas/kit#ncAA_Table The full list of amino acids used in this study can be found here]. |
| Line 108: | Line 108: | ||
| - | An ncAA synthetase/tRNA pair was cloned into pBLG and transformed into pFRYC amberless ''E. coli''. and pFRY amberless ''E. coli''. Other necessary control strains include RFP amberless ''E. coli'' (RFP control), sfGFP amberless ''E. coli'' (GFP control), amberless ''E. coli'' (cell background control), and LB media supplemented with ncAA (media background control). An overnight culture of each strain was grown in LB with the appropriate antibiotics at 37ºC and 225rpm. 10 mL of media with the appropriate antibiotics was inoculated with 100 µL of overnight culture and allowed to grow in the same conditions until the culture density was ~0.2-0.3 OD, or ~3 hours. The 10 mL culture was split between 4 different sterile test-tubes, 2 mL of culture per tube. The conditions of test tubes | + | An ncAA synthetase/tRNA pair was cloned into pBLG and transformed into pFRYC amberless ''E. coli''. and pFRY amberless ''E. coli''. Other necessary control strains include RFP amberless ''E. coli'' (RFP control), sfGFP amberless ''E. coli'' (GFP control), amberless ''E. coli'' (cell background control), and LB media supplemented with ncAA (media background control). An overnight culture of each strain was grown in LB with the appropriate antibiotics at 37ºC and 225rpm. 10 mL of media with the appropriate antibiotics was inoculated with 100 µL of overnight culture and allowed to grow in the same conditions until the culture density was ~0.2-0.3 OD, or ~3 hours. The 10 mL culture was split between 4 different sterile test-tubes, 2 mL of culture per tube. The conditions of the four test tubes were as follows: |
| + | *-IPTG,-ncAA | ||
| + | *-IPTG,+ncAA | ||
| + | *+IPTG, -ncAA | ||
| + | *+IPTG, +ncAA | ||
| + | IPTG stock solution was made at 1000X concentration ('''why is this here?''') and the ncAA was added to yield a concentration of 1 mM. Sterile deionized water was added in the place of ncAA and IPTG as a control ('''?'''). Once the controls, IPTG, and the ncAA were added appropriately, the cultures were allowed to grow to ~0.5 OD. 70 µL of each culture condition and control culture was added to a separate wells in a transparent 96-well plate for fluorescence and OD readings in a microplate reader. | ||
Revision as of 09:08, 14 October 2014
|
 "
"