Team:Austin Texas
From 2014.igem.org
| Line 79: | Line 79: | ||
<h1>Project: Expanded Genetic Code Measurement Kit</h1> | <h1>Project: Expanded Genetic Code Measurement Kit</h1> | ||
| - | Noncanonical amino acids (ncAAs) are an exciting new tool in the | + | Noncanonical amino acids (ncAAs) are an exciting new tool in the synthetic biologist's toolbox. Incorporating ncAAs into proteins will allow scientists to create bacteria that can perform novel functions. Unfortunately, these noncanonical amino acids are often difficult to effectively employ due to either low fidelity of the synthetase/tRNA pair or due to the pair having low efficiency. Our project aims to create a cheap, easy-to-use kit that can measure the fidelity and incorporation efficiency of numerous synthetase/tRNA pairs. The kit is a simple two-plasmid system. The first plasmid contains an IPTG-inducible RFP-sfGFP reporter. The two domains are connected by a linker sequence containing either a tyrosine codon (as a control) or a recoded amber stop codon (where the ncAA will be incorporated). The other plasmid will contain the synthetase/tRNA pair. After the RFP-linker-GFP protein is expressed, the fluorescence of each fluorescent domain of the fusion protein can be measured and compared. Depending on the relative intensities of the RFP and GFP fluorescence, we can determine both the fidelity of the synthetase/tRNA pair as well as the efficiency of the pair at incorporating the ncAA. We plan to equip researchers with a standardized, quick and easy “plug and play” system. Researchers will be able to insert any plasmid containing a new synthetase/tRNA pair into our Expanded Genetic Code Measurement kit to quickly characterize the synthetase/tRNA pair. |
| + | |||
|- valign="top" | |- valign="top" | ||
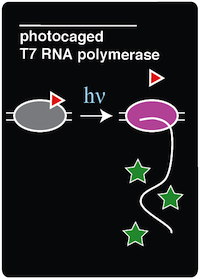
| [[Image:Austin_Texas_Photocage_Card.png|link=Team:Austin_Texas/photocage]] | | [[Image:Austin_Texas_Photocage_Card.png|link=Team:Austin_Texas/photocage]] | ||
Revision as of 14:21, 17 October 2014
| |||||||||||||||||||||||||||||||||||
 "
"